Diversity and community composition of methanogenic archaea in the rumen of Scottish upland sheep assessed by different methods - PubMed (original) (raw)
Diversity and community composition of methanogenic archaea in the rumen of Scottish upland sheep assessed by different methods
Timothy J Snelling et al. PLoS One. 2014.
Abstract
Ruminal archaeomes of two mature sheep grazing in the Scottish uplands were analysed by different sequencing and analysis methods in order to compare the apparent archaeal communities. All methods revealed that the majority of methanogens belonged to the Methanobacteriales order containing the Methanobrevibacter, Methanosphaera and Methanobacteria genera. Sanger sequenced 1.3 kb 16S rRNA gene amplicons identified the main species of Methanobrevibacter present to be a SGMT Clade member Mbb. millerae (≥ 91% of OTUs); Methanosphaera comprised the remainder of the OTUs. The primers did not amplify ruminal Thermoplasmatales-related 16S rRNA genes. Illumina sequenced V6-V8 16S rRNA gene amplicons identified similar Methanobrevibacter spp. and Methanosphaera clades and also identified the Thermoplasmatales-related order as 13% of total archaea. Unusually, both methods concluded that Mbb. ruminantium and relatives from the same clade (RO) were almost absent. Sequences mapping to rumen 16S rRNA and mcrA gene references were extracted from Illumina metagenome data. Mapping of the metagenome data to 16S rRNA gene references produced taxonomic identification to Order level including 2-3% Thermoplasmatales, but was unable to discriminate to species level. Mapping of the metagenome data to mcrA gene references resolved 69% to unclassified Methanobacteriales. Only 30% of sequences were assigned to species level clades: of the sequences assigned to Methanobrevibacter, most mapped to SGMT (16%) and RO (10%) clades. The Sanger 16S amplicon and Illumina metagenome mcrA analyses showed similar species richness (Chao1 Index 19-35), while Illumina metagenome and amplicon 16S rRNA analysis gave lower richness estimates (10-18). The values of the Shannon Index were low in all methods, indicating low richness and uneven species distribution. Thus, although much information may be extracted from the other methods, Illumina amplicon sequencing of the V6-V8 16S rRNA gene would be the method of choice for studying rumen archaeal communities.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
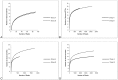
Figure 1. Multiple rarefaction collectors curves.
Observed number of OTUs for different sequencing and analysis methods: A. SA rrn. B. IM rrn. C. IM mcrA. D. IA rrn.
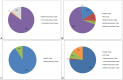
Figure 2. Relative distribution of methanogenic archaeal clades for different sequencing and analysis methods: A. SA rrn. B. IM rrn. C. IM mcrA. D. IA rrn.
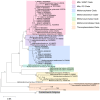
Figure 3. Phylogenetic analysis of SA rrn OTUs (RINH01–RINH21) and IA rrn OTUs (T01–T10).
Placement of representative sequences of the present study in clades indicated with additional reference sequences obtained from GenBank. A sequence related to the Crenarchaeota phylum (Acc. No. AF418935) was used as an outgroup. Full multiple alignment using ClustalW and a consensus tree was constructed using the Neighbor-Joining method with the Jukes-Cantor substitution model. The trees were bootstrap resampled 1000 times with branch values ≥50% shown. Scale shows 0.05 nucleotide substitutions per nucleotide position.
Similar articles
- Community structure of the metabolically active rumen bacterial and archaeal communities of dairy cows over the transition period.
Zhu Z, Noel SJ, Difford GF, Al-Soud WA, Brejnrod A, Sørensen SJ, Lassen J, Løvendahl P, Højberg O. Zhu Z, et al. PLoS One. 2017 Nov 8;12(11):e0187858. doi: 10.1371/journal.pone.0187858. eCollection 2017. PLoS One. 2017. PMID: 29117259 Free PMC article. - Structures of free-living and protozoa-associated methanogen communities in the bovine rumen differ according to comparative analysis of 16S rRNA and mcrA genes.
Tymensen LD, Beauchemin KA, McAllister TA. Tymensen LD, et al. Microbiology (Reading). 2012 Jul;158(Pt 7):1808-1817. doi: 10.1099/mic.0.057984-0. Epub 2012 Apr 26. Microbiology (Reading). 2012. PMID: 22539164 - Few highly abundant operational taxonomic units dominate within rumen methanogenic archaeal species in New Zealand sheep and cattle.
Seedorf H, Kittelmann S, Janssen PH. Seedorf H, et al. Appl Environ Microbiol. 2015 Feb;81(3):986-95. doi: 10.1128/AEM.03018-14. Epub 2014 Nov 21. Appl Environ Microbiol. 2015. PMID: 25416771 Free PMC article. - Molecular diversity of the rumen microbiome of Norwegian reindeer on natural summer pasture.
Sundset MA, Edwards JE, Cheng YF, Senosiain RS, Fraile MN, Northwood KS, Praesteng KE, Glad T, Mathiesen SD, Wright AD. Sundset MA, et al. Microb Ecol. 2009 Feb;57(2):335-48. doi: 10.1007/s00248-008-9414-7. Epub 2008 Jul 8. Microb Ecol. 2009. PMID: 18604648 - The 16S rRNA and mcrA gene based comparative diversity of methanogens in cattle fed on high fibre based diet.
Sirohi SK, Chaudhary PP, Singh N, Singh D, Puniya AK. Sirohi SK, et al. Gene. 2013 Jul 10;523(2):161-6. doi: 10.1016/j.gene.2013.04.002. Epub 2013 Apr 18. Gene. 2013. PMID: 23603353
Cited by
- Comparative analysis of rumen metagenome, metatranscriptome, fermentation and methane yield in cattle and buffaloes fed on the same diet.
Malik PK, Trivedi S, Kolte AP, Mohapatra A, Biswas S, Bhattar AVK, Bhatta R, Rahman H. Malik PK, et al. Front Microbiol. 2023 Nov 9;14:1266025. doi: 10.3389/fmicb.2023.1266025. eCollection 2023. Front Microbiol. 2023. PMID: 38029196 Free PMC article. - Evolving understanding of rumen methanogen ecophysiology.
Khairunisa BH, Heryakusuma C, Ike K, Mukhopadhyay B, Susanti D. Khairunisa BH, et al. Front Microbiol. 2023 Nov 6;14:1296008. doi: 10.3389/fmicb.2023.1296008. eCollection 2023. Front Microbiol. 2023. PMID: 38029083 Free PMC article. Review. - Holistic View and Novel Perspective on Ruminal and Extra-Gastrointestinal Methanogens in Cattle.
Aryee G, Luecke SM, Dahlen CR, Swanson KC, Amat S. Aryee G, et al. Microorganisms. 2023 Nov 10;11(11):2746. doi: 10.3390/microorganisms11112746. Microorganisms. 2023. PMID: 38004757 Free PMC article. Review. - Rumen microbial degradation of bromoform from red seaweed (Asparagopsis taxiformis) and the impact on rumen fermentation and methanogenic archaea.
Romero P, Belanche A, Jiménez E, Hueso R, Ramos-Morales E, Salwen JK, Kebreab E, Yáñez-Ruiz DR. Romero P, et al. J Anim Sci Biotechnol. 2023 Nov 1;14(1):133. doi: 10.1186/s40104-023-00935-z. J Anim Sci Biotechnol. 2023. PMID: 37907951 Free PMC article. - Diversity of rumen microbiota using metagenome sequencing and methane yield in Indian sheep fed on straw and concentrate diet.
Malik PK, Trivedi S, Kolte AP, Sejian V, Bhatta R, Rahman H. Malik PK, et al. Saudi J Biol Sci. 2022 Aug;29(8):103345. doi: 10.1016/j.sjbs.2022.103345. Epub 2022 Jun 16. Saudi J Biol Sci. 2022. PMID: 35770269 Free PMC article.
References
- Cavicchioli R (2011) Archaea - timeline of the third domain. Nature Rev Microbiol 9: 51–61. - PubMed
- Czerkawski JW (1969) Methane production in ruminants and its significance. World Rev Nutr Diet 11: 240–282. - PubMed
- Moss AR, Jouany JP, Newbold J (2000) Methane production by ruminants: Its contribution to global warming. Ann Zootech 49: 231–253.
- Scottish government (2010) Abstract of Scottish agricultural statistics 1982 to 2009. Available: http://www.scotland.gov.uk/Publications/2010/03/16160036/24. Accessed 2014 March 17.
- Scottish government (2009) Climate change (Scotland) Act 2009 (asp 12). Available: www.legislation.gov.uk/asp/2009/12/contents. Accessed 2014 March 17.
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/D/20211551/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004413/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004243/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004235/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211553/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous