Discrepancies in cancer genomic sequencing highlight opportunities for driver mutation discovery - PubMed (original) (raw)
Discrepancies in cancer genomic sequencing highlight opportunities for driver mutation discovery
Andrew M Hudson et al. Cancer Res. 2014.
Abstract
Cancer genome sequencing is being used at an increasing rate to identify actionable driver mutations that can inform therapeutic intervention strategies. A comparison of two of the most prominent cancer genome sequencing databases from different institutes (Cancer Cell Line Encyclopedia and Catalogue of Somatic Mutations in Cancer) revealed marked discrepancies in the detection of missense mutations in identical cell lines (57.38% conformity). The main reason for this discrepancy is inadequate sequencing of GC-rich areas of the exome. We have therefore mapped over 400 regions of consistent inadequate sequencing (cold-spots) in known cancer-causing genes and kinases, in 368 of which neither institute finds mutations. We demonstrate, using a newly identified PAK4 mutation as proof of principle, that specific targeting and sequencing of these GC-rich cold-spot regions can lead to the identification of novel driver mutations in known tumor suppressors and oncogenes. We highlight that cross-referencing between genomic databases is required to comprehensively assess genomic alterations in commonly used cell lines and that there are still significant opportunities to identify novel drivers of tumorigenesis in poorly sequenced areas of the exome. Finally, we assess other reasons for the observed discrepancy, such as variations in dbSNP filtering and the acquisition/loss of mutations, to give explanations as to why there is a discrepancy in pharmacogenomic studies, given recent concerns with poor reproducibility of data.
©2014 American Association for Cancer Research.
Figures
Figure 1
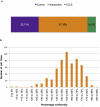
Marked discrepancy is seen in mutation calling between CCLE and COSMIC. a) Overall percentage conformity of 46,409 mutations detected by COSMIC and/or CCLE. The intersection between datasets (mutations found by both institutes) accounted for 57.38%. Cosmic-only mutations comprised 32.71% of the dataset and CCLE only mutations 9.91%. b) The percentage agreement between mutations reported in the 568 cell lines sequenced by both institutes.
Figure 2
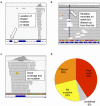
In the original 18 cell line comparison, mutations detected by COSMIC but not CCLE were categorised into: poor coverage with 5 or less reads (Panel a); good read coverage (over 20 reads) and mutation detected on reads but annotated as a dbSNP, neutral variant, outside coding region in all transcripts, or detected on less than 10% of reads, and removed (Panel b); and good coverage, no mutation (Panel c). (Panel d) reveals that the most common cause for mutations being missed by CCLE was poor read coverage (41%). Images of read coverage were taken using the Integrative Genomics Viewer.
Figure 3
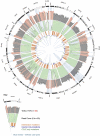
The 20 largest cold-spots detected in cancer census or kinase genes transcripts (of those that were sequenced by both COSMIC and CCLE hybrid capture) using CCLE whole exome sequencing data. All but one of these cold-spots is located in a high GC-content area and results in no mutations being detected by either institute. The TET2 cold-spot is not located in high-GC content areas and contains mutations detected by COSMIC, indicating that this cold-spot was not present in the COSMIC data. The outer shaded grey plot shows the GC content at each base (calculated as 50bp either side) with GC content over 70% shaded in red. The middle light green plot shows sequencing read coverage with white troughs representing poor read coverage. The inner 3 rings record the position of mutations found by both institutes (orange), COSMIC only (violet) and CCLE (green). Light blue shards show cold-spots over 100bp in length with the top 20 shaded darker. Data were plotted using a combination of Circos and custom scripts.
Similar articles
- Exome-wide Sequencing Shows Low Mutation Rates and Identifies Novel Mutated Genes in Seminomas.
Cutcutache I, Suzuki Y, Tan IB, Ramgopal S, Zhang S, Ramnarayanan K, Gan A, Lee HH, Tay ST, Ooi A, Ong CK, Bolthouse JT, Lane BR, Anema JG, Kahnoski RJ, Tan P, Teh BT, Rozen SG. Cutcutache I, et al. Eur Urol. 2015 Jul;68(1):77-83. doi: 10.1016/j.eururo.2014.12.040. Epub 2015 Jan 14. Eur Urol. 2015. PMID: 25597018 - Exome sequencing reveals comprehensive genomic alterations across eight cancer cell lines.
Chang H, Jackson DG, Kayne PS, Ross-Macdonald PB, Ryseck RP, Siemers NO. Chang H, et al. PLoS One. 2011;6(6):e21097. doi: 10.1371/journal.pone.0021097. Epub 2011 Jun 20. PLoS One. 2011. PMID: 21701589 Free PMC article. - [Lung cancer molecular testing, what role for Next Generation Sequencing and circulating tumor DNA].
Pécuchet N, Legras A, Laurent-Puig P, Blons H. Pécuchet N, et al. Ann Pathol. 2016 Jan;36(1):80-93. doi: 10.1016/j.annpat.2015.11.012. Epub 2016 Jan 20. Ann Pathol. 2016. PMID: 26803564 French. - Sequencing approaches in cancer treatment.
Sekar D, Thirugnanasambantham K, Hairul Islam VI, Saravanan S. Sekar D, et al. Cell Prolif. 2014 Oct;47(5):391-5. doi: 10.1111/cpr.12124. Epub 2014 Aug 7. Cell Prolif. 2014. PMID: 25131793 Free PMC article. Review. - Understanding oncogenicity of cancer driver genes and mutations in the cancer genomics era.
Porta-Pardo E, Valencia A, Godzik A. Porta-Pardo E, et al. FEBS Lett. 2020 Dec;594(24):4233-4246. doi: 10.1002/1873-3468.13781. Epub 2020 Apr 28. FEBS Lett. 2020. PMID: 32239503 Free PMC article. Review.
Cited by
- HuH-7 reference genome profile: complex karyotype composed of massive loss of heterozygosity.
Kasai F, Hirayama N, Ozawa M, Satoh M, Kohara A. Kasai F, et al. Hum Cell. 2018 Jul;31(3):261-267. doi: 10.1007/s13577-018-0212-3. Epub 2018 May 17. Hum Cell. 2018. PMID: 29774518 Free PMC article. - COSMIC: exploring the world's knowledge of somatic mutations in human cancer.
Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M, De T, Teague JW, Stratton MR, McDermott U, Campbell PJ. Forbes SA, et al. Nucleic Acids Res. 2015 Jan;43(Database issue):D805-11. doi: 10.1093/nar/gku1075. Epub 2014 Oct 29. Nucleic Acids Res. 2015. PMID: 25355519 Free PMC article. - ApoCanD: Database of human apoptotic proteins in the context of cancer.
Kumar R, Raghava GP. Kumar R, et al. Sci Rep. 2016 Feb 10;6:20797. doi: 10.1038/srep20797. Sci Rep. 2016. PMID: 26861916 Free PMC article. - Identifying CpG sites with different differential methylation frequencies in colorectal cancer tissues based on individualized differential methylation analysis.
Yan H, He J, Guan Q, Cai H, Zhang L, Zheng W, Qi L, Zhang S, Liu H, Li H, Zhao W, Yang S, Guo Z. Yan H, et al. Oncotarget. 2017 Jul 18;8(29):47356-47364. doi: 10.18632/oncotarget.17647. Oncotarget. 2017. PMID: 28537885 Free PMC article. - Evaluating the consistency of large-scale pharmacogenomic studies.
Rahman R, Dhruba SR, Matlock K, De-Niz C, Ghosh S, Pal R. Rahman R, et al. Brief Bioinform. 2019 Sep 27;20(5):1734-1753. doi: 10.1093/bib/bby046. Brief Bioinform. 2019. PMID: 31846027 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- 15675/CRUK_/Cancer Research UK/United Kingdom
- A3669/CRUK_/Cancer Research UK/United Kingdom
- 12936/CRUK_/Cancer Research UK/United Kingdom
- A15675/CRUK_/Cancer Research UK/United Kingdom
- 17098/CRUK_/Cancer Research UK/United Kingdom
- 17263/CRUK_/Cancer Research UK/United Kingdom
- A12936/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous