RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites - PubMed (original) (raw)
RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites
Jesse M Engreitz et al. Cell. 2014.
Abstract
Intermolecular RNA-RNA interactions are used by many noncoding RNAs (ncRNAs) to achieve their diverse functions. To identify these contacts, we developed a method based on RNA antisense purification to systematically map RNA-RNA interactions (RAP-RNA) and applied it to investigate two ncRNAs implicated in RNA processing: U1 small nuclear RNA, a component of the spliceosome, and Malat1, a large ncRNA that localizes to nuclear speckles. U1 and Malat1 interact with nascent transcripts through distinct targeting mechanisms. Using differential crosslinking, we confirmed that U1 directly hybridizes to 5' splice sites and 5' splice site motifs throughout introns and found that Malat1 interacts with pre-mRNAs indirectly through protein intermediates. Interactions with nascent pre-mRNAs cause U1 and Malat1 to localize proximally to chromatin at active genes, demonstrating that ncRNAs can use RNA-RNA interactions to target specific pre-mRNAs and genomic sites. RAP-RNA is sensitive to lower abundance RNAs as well, making it generally applicable for investigating ncRNAs.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
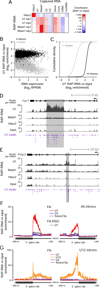
Figure 1. U1 Binds Throughout Nascent Transcripts at 5’-Splice-Site Motifs
(A) Schematic diagram of U1 RAP-RNA[AMT]. AMT forms covalent crosslinks (orange) between opposing uridine bases in the U1 snRNA (red) and target pre-mRNA (gray). Consensus sequences show canonical base-pairing interaction. To avoid capturing indirectly associated RNAs, we digested protein and DNA before purifying U1 with biotinylated antisense probes (blue). To map RNA-RNA interaction sites at high resolution, we fragmented RNA prior to capture and performed reverse transcription (RT) without reversal of crosslinks, leading to complementary DNA (cDNA, purple) that terminates at or near the site of a crosslink (see also Figure S1). Ligation of a second adapter to the 3’ end of the cDNA enabled sequencing and mapping the positions of RT termination. (B) Read counts aggregated over all 5’ splice sites. Each read-pair contributes a count in the base corresponding to the 5’ end of the original RNA fragment. Cells were crosslinked with AMT (+AMT, red) or mock-crosslinked with DMSO (−AMT, blue). Crosslinked input RNA (black) is shown for comparison. (C) Enrichment for every 8-mer RNA motif close to RAP-RNA sequencing reads. Colored dots represent 8-mers that are significantly enriched in U1 RAP-RNA versus input (P < 0.001 after Bonferroni correction, enrichment >= 4). Red, orange, and yellow dots correspond to 8-mers that have a Levenshtein edit distance from the 8-mer consensus motif of 1, 2, or >2, respectively. (D) Enrichment for sequencing reads in U1 RAP-RNA[AMT] versus input at 5’ splice sites and at 5’ss motif matches >200 bases away from exons. Enrichments are normalized to the enrichment at random intronic sites >200 bases away from exons. 5’ss motif matches were classified as strong, medium, or weak as previously described (Almada et al., 2013). Strong AS = strong motifs on the antisense strand, which should not be bound by U1. (E) Same as (C), but considering only reads that map to introns >200 bases away from exons. (F) U1 RAP-RNA[AMT] coverage and enrichment across the Malat1 transcript (10-nt resolution), representing the ratio of U1 RAP +AMT and the maximum of U1 RAP −AMT and Input +AMT. Black bars represent significantly enriched windows (P < 0.001 after Bonferroni correction). Conservation represents the phyloP30wayPlacental track from the UCSC Genome Browser. (G) and (H) Zoom-in on two significant U1 binding sites. Scales on _y_-axes are the same as in (F).
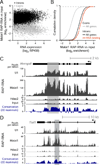
Figure 2. RAP-RNA Captures Both Direct and Indirect RNA-RNA Interactions
(A) Enrichment for individual transcripts or classes of RNAs. RAP-RNA[FA-DSG] experiments include two replicate U1 and Malat1 purifications (rep1 and rep2) and one Hdac2 purification (negative control). (B) Each point represents the average U1 RAP-RNA[FA-DSG] enrichment across all introns (black) or exons (gray) for one gene. The _x_-axis represents the expression of the mature transcript (exons) in input nuclear-enriched RNA in reads per kilobase per million (RPKM). (C) Comparison of U1 RAP-RNA[FA-DSG] enrichment for the exons (gray) and introns (black) of all genes. (D) RAP-RNA[FA-DSG] sequencing reads mapping to Gpi1 and (E) Ptbp3, two of the most highly enriched transcripts in U1 RAP-RNA[FA-DSG] (>100-fold versus input). Gray rectangles highlight enriched introns with many U1 motifs (purple). (F) Enrichment of sequencing reads in RAP-RNA[FA] versus input aggregated across 5’ and 3’ splice sites for all introns (n > 180,000) or (G) U12 introns (n = 588). U1 RAP-RNA[FA] (red) more strongly enriches splice sites than U1 RAP-RNA[FA-DSG] (gray). Each read-pair contributes a count at the position corresponding to the 3’ end of the original RNA fragment for the 5’-splice-site panel and the 5’ end for the 3’-splice-site panel. See also Figure S2.
Figure 3. Malat1 Interacts Indirectly with Nascent Transcripts Encoding RNA-Binding Proteins
(A) Malat1 RAP-RNA[FA-DSG] enrichment at exons and introns, similar to Figure 2B. (B) Comparison of Malat1 RAP-RNA[FA-DSG] enrichment for the exons (gray) and introns (black) of all genes or of significant gene sets. See also Figure S3. (C) Malat1 RAP-RNA[FA-DSG] sequencing reads mapping to Hnrnpdl and (D) Tial1, two highly enriched genes encoding RNA-binding proteins. Gray rectangles highlight conserved introns containing RNA elements that control alternative splicing.
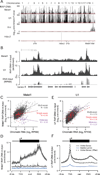
Figure 4. Chromatin Localization of U1 and Malat1
(A) Significance of enrichment across the genome in 10-kb windows for Malat1, U1, Xist, and Hdac2 RAP-DNA. _Y_-axis represents uncorrected binomial _P_-values, and the red lines mark the cutoff for genome-wide significance (P < 10−10). Ticks at the bottom denote the locations of the genes encoding each RNA. U1 is encoded on chromosomes 3, 11, and 12. (B) RAP-DNA enrichment versus input across a region containing several active genes (chr18: 34,313,699–35,191,532). RNA sequencing of input total RNA shows relative gene expression. (C) Scatterplot shows Malat1 RAP-DNA enrichment versus levels of chromatin-associated RNA for single-exon histone genes (purple), single-exon non-histone genes (red), and all other genes (dark gray). Dashed lines represent linear regressions. R = Pearson’s correlation. (D) RAP-DNA enrichment averaged over active genes (black), inactive genes (blue), and active genes in cells treated with flavopiridol (black dashed line). Shaded regions represent 95% confidence intervals for the average enrichment. For Malat1, the averages represent the 5% of active genes with the highest Malat1 enrichment and an equal number of randomly selected inactive genes. (E) Same as (C) for U1 RAP-DNA. (F) Same as (D) for U1 RAP-DNA. For U1, average enrichments include all active and inactive genes. Notable peaks occur at the transcription start site (TSS) and at the polyadenylation site (PAS). See also Figure S4.
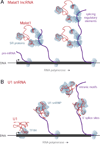
Figure 5. Malat1 and U1 Interact with Pre-mRNAs and Chromatin Through Different Mechanisms
(A) Malat1 interacts indirectly with nascent RNAs at splicing regulatory elements, like the conserved noncoding elements in Hnrnpdl and Tial1, possibly through its known interactions with SR splicing proteins (Tripathi et al., 2010). These interactions enable Malat1 to localize to chromatin at active gene loci. Malat1 RAP may co-purify DNA through tethering of the nascent transcript to chromatin by RNA polymerase. (B) U1 interacts with pre-mRNAs through direct hybridization, both at 5’ splice sites and throughout introns. U1 interacts with chromatin through two different mechanisms: (i) U1 localizes throughout active gene loci as a secondary result of its direct interactions with the nascent pre-mRNA, and (ii) U1 localizes at the 5’ ends of genes through a mechanism that does not depend on transcriptional elongation, perhaps via known interactions with the cyclin H subunit of TFIIH (Kwek et al., 2002). While these figures depict interactions with chromatin-associated pre-mRNAs, U1 and Malat1 may also interact with pre-mRNAs after their release from chromatin. snRNP: small nuclear ribonucleic particle.
Comment in
- Non-coding RNA: a match made by crosslinking.
Lau E. Lau E. Nat Rev Genet. 2014 Nov;15(11):704. doi: 10.1038/nrg3842. Epub 2014 Oct 9. Nat Rev Genet. 2014. PMID: 25297729 No abstract available.
Similar articles
- U1 snRNP regulates chromatin retention of noncoding RNAs.
Yin Y, Lu JY, Zhang X, Shao W, Xu Y, Li P, Hong Y, Cui L, Shan G, Tian B, Zhang QC, Shen X. Yin Y, et al. Nature. 2020 Apr;580(7801):147-150. doi: 10.1038/s41586-020-2105-3. Epub 2020 Mar 11. Nature. 2020. PMID: 32238924 - Evolutionarily divergent spliceosomal snRNAs and a conserved non-coding RNA processing motif in Giardia lamblia.
Hudson AJ, Moore AN, Elniski D, Joseph J, Yee J, Russell AG. Hudson AJ, et al. Nucleic Acids Res. 2012 Nov;40(21):10995-1008. doi: 10.1093/nar/gks887. Epub 2012 Sep 27. Nucleic Acids Res. 2012. PMID: 23019220 Free PMC article. - The U1 snRNP protein U1C recognizes the 5' splice site in the absence of base pairing.
Du H, Rosbash M. Du H, et al. Nature. 2002 Sep 5;419(6902):86-90. doi: 10.1038/nature00947. Nature. 2002. PMID: 12214237 - RNA motifs and combinatorial prediction of interactions, stability and localization of noncoding RNAs.
Gandhi M, Caudron-Herger M, Diederichs S. Gandhi M, et al. Nat Struct Mol Biol. 2018 Dec;25(12):1070-1076. doi: 10.1038/s41594-018-0155-0. Epub 2018 Nov 12. Nat Struct Mol Biol. 2018. PMID: 30420773 Review. - U1 snRNP Telescripting: Suppression of Premature Transcription Termination in Introns as a New Layer of Gene Regulation.
Venters CC, Oh JM, Di C, So BR, Dreyfuss G. Venters CC, et al. Cold Spring Harb Perspect Biol. 2019 Feb 1;11(2):a032235. doi: 10.1101/cshperspect.a032235. Cold Spring Harb Perspect Biol. 2019. PMID: 30709878 Free PMC article. Review.
Cited by
- Advances in endogenous RNA pull-down: A straightforward dextran sulfate-based method enhancing RNA recovery.
Desideri F, D'Ambra E, Laneve P, Ballarino M. Desideri F, et al. Front Mol Biosci. 2022 Oct 19;9:1004746. doi: 10.3389/fmolb.2022.1004746. eCollection 2022. Front Mol Biosci. 2022. PMID: 36339717 Free PMC article. - LncRNAs in vertebrates: advances and challenges.
Mallory AC, Shkumatava A. Mallory AC, et al. Biochimie. 2015 Oct;117:3-14. doi: 10.1016/j.biochi.2015.03.014. Epub 2015 Mar 24. Biochimie. 2015. PMID: 25812751 Free PMC article. Review. - _ESR1_-Stabilizing Long Noncoding RNA TMPO-AS1 Promotes Hormone-Refractory Breast Cancer Progression.
Mitobe Y, Ikeda K, Suzuki T, Takagi K, Kawabata H, Horie-Inoue K, Inoue S. Mitobe Y, et al. Mol Cell Biol. 2019 Nov 12;39(23):e00261-19. doi: 10.1128/MCB.00261-19. Print 2019 Dec 1. Mol Cell Biol. 2019. PMID: 31501276 Free PMC article. - PRAS40 hyperexpression promotes hepatocarcinogenesis.
Qi Z, Zhang T, Song L, Fu H, Luo H, Wu J, Zhao S, Zhang T, Guo L, Jin L, Zhang H, Huang G, Ma T, Wu Y, Huang L. Qi Z, et al. EBioMedicine. 2020 Jan;51:102604. doi: 10.1016/j.ebiom.2019.102604. Epub 2020 Jan 3. EBioMedicine. 2020. PMID: 31901857 Free PMC article. - Functional diversity of small nucleolar RNAs.
Bratkovič T, Božič J, Rogelj B. Bratkovič T, et al. Nucleic Acids Res. 2020 Feb 28;48(4):1627-1651. doi: 10.1093/nar/gkz1140. Nucleic Acids Res. 2020. PMID: 31828325 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases