Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials - PubMed (original) (raw)
. 2014 Nov;32(11):1146-50.
doi: 10.1038/nbt.3043. Epub 2014 Oct 5.
Affiliations
- PMID: 25282355
- PMCID: PMC4317352
- DOI: 10.1038/nbt.3043
Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials
David Bikard et al. Nat Biotechnol. 2014 Nov.
Abstract
Antibiotics target conserved bacterial cellular pathways or growth functions and therefore cannot selectively kill specific members of a complex microbial population. Here, we develop programmable, sequence-specific antimicrobials using the RNA-guided nuclease Cas9 (refs.1,2) delivered by a bacteriophage. We show that Cas9, reprogrammed to target virulence genes, kills virulent, but not avirulent, Staphylococcus aureus. Reprogramming the nuclease to target antibiotic resistance genes destroys staphylococcal plasmids that harbor antibiotic resistance genes and immunizes avirulent staphylococci to prevent the spread of plasmid-borne resistance genes. We also show that CRISPR-Cas9 antimicrobials function in vivo to kill S. aureus in a mouse skin colonization model. This technology creates opportunities to manipulate complex bacterial populations in a sequence-specific manner.
Figures
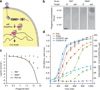
Figure 1
Sequence-specific killing of S. aureus by a phagemid-delivered CRISPR system. (a) The ΦNM1 phage delivers the pDB121 phagemid to S. aureus cells. pDB121 carries the S. pyogenes tracrRNA, cas9 and a programmable CRISPR array sequence. Expression of cas9 and a self-targeting crRNA leads to chromosome cleavage and cell death. (b) Lysates of pDB121 phagemid targeting the aph-3 kanamycin resistance gene or a non-targeting control are spotted on top-agar lawns of either RNΦ or RNKΦ cells. Scale bar, 5mm (c) Treatment of RNΦ (blue triangles) or RNKΦ (red diamonds) with pDB121::aph at various MOI. Survival is calculated as the ratio of CFU recovered after treatment to CFU from an untreated sample of the same culture (mean ± s.d.). The black curve represents the probability that a cell does not receive a phagemid making the assumption that all cells have the same chance of receiving phagemid. (d) Time course of treatment of RNKΦ/pCN57 (GFP reporter plasmid) cells in a mixed culture with non-targeted RNΦ cells. Plain lines show OD and dashed lines GFP. Kanamycin (25 ug/ml) is shown in orange, streptomycin (10 ug/ml) in green, pDB121::aph (MOI ~20) in purple, pDB121::Ø (MOI ~20) in blue.
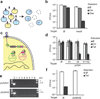
Figure 2
Targeting antibiotic resistance genes and plasmids in an MRSA strain. (a) Treatment of a mixed population of RNΦ and USA300Φ results in killing of the targeted USA300 MRSA strain and delivery of an immunizing phagemid to the rest of the population. (b) pDB121::mecA specifically kills USA300Φ in a mixed population. Exponentially growing USA300Φ and RNΦ cells were mixed 1:1 and treated with pDB121 at an MOI of ~5. Cells were plated either on a non-selective medium, on chloramphenicol-containing medium to measure the proportion of cells receiving the phagemid treatment, or on oxacillin-containing medium to measure the proportion of USA300Φ cells in the population (mean ± s.d.). (c) The CRISPR array sequence is programmed to target the pUSA01 and pUSA02 plasmids simultaneously. (d) USA300Φ was treated with pDB121 lysates targeting each plasmid individually or in combination. Cells were plated either on a non-selective medium, on chloramphenicol-containing medium to measure the proportion of cells receiving the phagemid treatment, or on tetracycline-containing medium to measure the proportion of cells cured of pUSA02 (mean ± s.d.). (e) Plasmid curing was confirmed by the lack of PCR amplification with plasmid specific oligonucleotides in 8 independent CFUs after treatment with the double targeting construct. (f) A population of RNΦ cells was immunized against plasmid horizontal transfer by treatment with the pUSA02-targeting pDB121 phagemid. 30 min after treatment, the population is transduced with a ΦNM1 stock grown on USA300. Cells are plated either without selection or on tetracycline to measure transduction efficiency of the pUSA02 plasmid (mean ± s.d.).
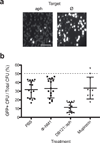
Figure 3
Sequence-specific killing of kanamycin resistant S. aureus in a mouse skin colonization model. Mice skin was colonized with a 1:1 mixture of 105 RNΦ and RNKΦ cells carrying the pCN57 GFP reporter plasmid, followed by treatment at 1 hour with phosphate buffer saline (n=15), ΦNM1 (n=16), pDB121::aph (n=14) or mupirocin (n=10). After 24 hours the skin from the treated area was excised, homogenized and serial dilutions of the homogenate was plated on mannitol salt agar (a) Pictures of two representative plates exposed to a wavelength enabling visualization of GFP. Scale bar, 1 cm. (b) The proportion of RNKΦ cells in the population was measured as the proportion of green fluorescent cfu on the plates. Data points indicate individual mice; black lines represent the mean ± SD.
Comment in
- A CRISPR design for next-generation antimicrobials.
Beisel CL, Gomaa AA, Barrangou R. Beisel CL, et al. Genome Biol. 2014 Nov 8;15(11):516. doi: 10.1186/s13059-014-0516-x. Genome Biol. 2014. PMID: 25417800 Free PMC article.
Similar articles
- Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases.
Citorik RJ, Mimee M, Lu TK. Citorik RJ, et al. Nat Biotechnol. 2014 Nov;32(11):1141-5. doi: 10.1038/nbt.3011. Epub 2014 Sep 21. Nat Biotechnol. 2014. PMID: 25240928 Free PMC article. - Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus.
Park JY, Moon BY, Park JW, Thornton JA, Park YH, Seo KS. Park JY, et al. Sci Rep. 2017 Mar 21;7:44929. doi: 10.1038/srep44929. Sci Rep. 2017. PMID: 28322317 Free PMC article. - A CRISPR design for next-generation antimicrobials.
Beisel CL, Gomaa AA, Barrangou R. Beisel CL, et al. Genome Biol. 2014 Nov 8;15(11):516. doi: 10.1186/s13059-014-0516-x. Genome Biol. 2014. PMID: 25417800 Free PMC article. - CRISPR-Cas-Based Antimicrobials: Design, Challenges, and Bacterial Mechanisms of Resistance.
Mayorga-Ramos A, Zúñiga-Miranda J, Carrera-Pacheco SE, Barba-Ostria C, Guamán LP. Mayorga-Ramos A, et al. ACS Infect Dis. 2023 Jul 14;9(7):1283-1302. doi: 10.1021/acsinfecdis.2c00649. Epub 2023 Jun 22. ACS Infect Dis. 2023. PMID: 37347230 Free PMC article. Review. - Using CRISPR-Cas systems as antimicrobials.
Bikard D, Barrangou R. Bikard D, et al. Curr Opin Microbiol. 2017 Jun;37:155-160. doi: 10.1016/j.mib.2017.08.005. Epub 2017 Sep 6. Curr Opin Microbiol. 2017. PMID: 28888103 Review.
Cited by
- Type III-A CRISPR immunity promotes mutagenesis of staphylococci.
Mo CY, Mathai J, Rostøl JT, Varble A, Banh DV, Marraffini LA. Mo CY, et al. Nature. 2021 Apr;592(7855):611-615. doi: 10.1038/s41586-021-03440-3. Epub 2021 Apr 7. Nature. 2021. PMID: 33828299 Free PMC article. - Insights from 20 years of bacterial genome sequencing.
Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW. Land M, et al. Funct Integr Genomics. 2015 Mar;15(2):141-61. doi: 10.1007/s10142-015-0433-4. Epub 2015 Feb 27. Funct Integr Genomics. 2015. PMID: 25722247 Free PMC article. Review. - In silico optimization of RNA-protein interactions for CRISPR-Cas13-based antimicrobials.
Park HM, Park Y, Berani U, Bang E, Vankerschaver J, Van Messem A, De Neve W, Shim H. Park HM, et al. Biol Direct. 2022 Oct 7;17(1):27. doi: 10.1186/s13062-022-00339-5. Biol Direct. 2022. PMID: 36207756 Free PMC article. - A bug's life: Delving into the challenges of helminth microbiome studies.
Formenti F, Cortés A, Brindley PJ, Cantacessi C, Rinaldi G. Formenti F, et al. PLoS Negl Trop Dis. 2020 Sep 10;14(9):e0008446. doi: 10.1371/journal.pntd.0008446. eCollection 2020 Sep. PLoS Negl Trop Dis. 2020. PMID: 32911483 Free PMC article. Review. No abstract available. - Potency of CRISPR-Cas Antifungals Is Enhanced by Cotargeting DNA Repair and Growth Regulatory Machinery at the Genetic Level.
Mendoza BJ, Zheng X, Clements JC, Cotter C, Trinh CT. Mendoza BJ, et al. ACS Infect Dis. 2023 Dec 8;9(12):2494-2503. doi: 10.1021/acsinfecdis.3c00342. Epub 2023 Nov 13. ACS Infect Dis. 2023. PMID: 37955405 Free PMC article.
References
- Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. - PubMed
- Weigel LM, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DP2 AI104556/AI/NIAID NIH HHS/United States
- R01 AI057472/AI/NIAID NIH HHS/United States
- T32 AI070084/AI/NIAID NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical