Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4 - PubMed (original) (raw)
Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4
Brant M Webster et al. Cell. 2014.
Abstract
The maintenance of nuclear compartmentalization by the nuclear envelope and nuclear pore complexes (NPCs) is essential for cell function; loss of compartmentalization is associated with cancers, laminopathies, and aging. We uncovered a pathway that surveils NPC assembly intermediates to promote the formation of functional NPCs. Surveillance is mediated by Heh2, a member of the LEM (Lap2-emerin-MAN1) family of integral inner nuclear membrane proteins, which binds to an early NPC assembly intermediate, but not to mature NPCs. Heh2 recruits the endosomal sorting complex required for transport (ESCRT)-III subunit Snf7 and the AAA-ATPase Vps4 to destabilize and clear defective NPC assembly intermediates. When surveillance or clearance is compromised, malformed NPCs accumulate in a storage of improperly assembled nuclear pore complexes compartment, or SINC. The SINC is retained in old mothers to prevent loss of daughter lifespan, highlighting a continuum of mechanisms to ensure nuclear compartmentalization.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
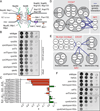
Figure 1. ESCRT-III/VPS4 specifically interact with nup genes
(A) Schematic of the NPC. Nups shown in the inner (red oval), outer (blue oval), and membrane (orange oval) ring complexes; nuclear basket and cytoplasmic filaments in green. FG-nups are grey lines and linker nup Nic96 is grey oval. Integral (Heh1/2) and associated (Esc1) INM proteins shown. (B) POM152 interacts with ESCRT-III genes and VPS4. (C) Quantification of epistasis (ε) between POM152 and ESCRT genes. ε can range from −1 to 1 where negative numbers are synthetic sick interactions and positive numbers are suppressive interactions. Mean +/- s.d. from 3 experiments. “E” abbreviates ESCRT. (D and E) Schematic of all tested genetic interactions. (F) VPS4 and SNF7 genetically interact with HEH2. See Figure S1.
Figure 2. Snf7 physically interacts with Heh1 and Heh2 at the NE
(A) Diagram of BifC where bait and prey are co-expressed as VN and VC fragments of Venus. Interactions of bait and prey result in Venus fluorescence. (B) Snf7 interacts with Heh1, Heh2 and Nup170 at the NE. Fluorescent micrographs of cells expressing VC and VN fusions. Background fluorescence has been enhanced to facilitate interpretation of the low signal to noise in some cells. Cell boundaries traced from transmitted light images (not shown). Bar is 5µm. Insets are a merge of fluorescent micrographs of Venus and the NE/ER marker HDEL-dsRed. In some cells vacuoles (V) are visible due to autofluorescence. Bar in inset is 1 µm. (C) Plot of the percentage of cells expressing VN and VC fusions where Venus was observed. Plots are mean +/− s.d. from 3 experiments (n>200). (D) Plot of the percentage of fluorescent spots seen colocalized with HDEL-dsRED at the NE. Mean +/− s.d. from 3 experiments (n>40). See Figure S2.
Figure 3. Heh2 specifically binds Snf7
(A) Snf7 co-purifies with Heh2-GFP in _vps4_Δ cells. Western blots of proteins bound to Heh1- and Heh2-GFP from WT and _vps4_Δ cell extracts. (B) Schematic of the topology and domain architecture of Heh2. LEM is Lap2-emerin-MAN1, MCHD is MAN1 C-terminal Homology Domain and TMD is transmembrane domain. (C and D) Snf7 specifically interacts with the N-terminus of Heh2. GST-fusions of Heh2 domains were immobilized on GST-beads (bottom, coomassie panels) and incubated with buffer or whole cell extracts (WCE) as indicated in legend. HA fusions detected by Western blot. See Figure S3.
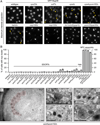
Figure 4. Nups accumulate in a cluster at the NE in the absence of ESCRT-III/Vps4
(A) GFP-Nup49 accumulates at one side of the NE in _snf7_Δ and _vps4_Δ strains. Maximum intensity projections shown. Arrowheads point to the nup cluster. Bar is 5µm. (B) Plot of the percentage of cells in the indicated strains where GFP-Nup49 accumulates in a cluster. Mean +/− s.d. from 3 experiments (n>400). (C-G) TEM micrographs of _vps4Δpom152_Δ cells showing an accumulation of NPC-like structures on one side of the NE (pseudo-colored red) in addition to intralumenal vesicles. All bars are 200 nm. In (C), an anti-Nsp1 antibody and 10nm-gold conjugated secondary antibodies label the NPC-like structures. “N” and “C” represent nucleoplasm and cytoplasm, respectively. Arrows, double arrows and arrowhead point to the ONM, INM and INM invaginations, respectively. Asterisks label putative vesicles in the NE lumen.
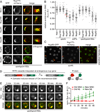
Figure 5. The SINC is a product of NPC misassembly
(A) The SINC contains an irregular nup composition depleted of cytoplasmic and nucleoplasmic nups. _vps4Δpom152_Δ cells expressing GFP-tagged nups and Nup170-mCherry. The 4th column is an orthogonal (x,z or y,z) view of the SINC. Bar is 1 µm. (B) Plot of the relative nup-GFP enrichment (Equery) in the SINC compared to Nup170-mCherry (Eref) expressed as a Equery:Eref ratio. Line is mean, error bars are s.d., grey circles are individual cells, and red circles with numbers correspond to the indicated cells in (C). n=20 in each of 3 experiments. (C) Variability in the relative accumulation of Nup60-GFP and Nup170-mCherry in the SINC. Arrowheads point to SINCs, numbers refer to individual cells in (B). Bar is 5µm. (D) Schematic of RITE where estradiol induces a genetic switch from the production of Nup-mCherry to Nup-GFP. EBD is estradiol binding domain. (E) Newly synthesized Nup85-GFP accumulates in the SINC. Fluorescent micrographs of the nucleus of a _vps4Δpom152_Δ cell showing Nup85-GFP (New/85-GFP), Nup85-mCherry (Old/85-mCh) and merged images at 10 min intervals in the presence of estradiol. Bar is 1 µm. (F) Quantification of Nup85-GFP and mCherry fluorescence (normalized fluorescence intensity in arbitrary units (A.U.)) at the NE and SINC after estradiol addition, values normalized to t=0. Mean +/− s.d. from 3 experiments (n=3, 5, 5). See Figure S4.
Figure 6. The SINC is not transmitted to daughter cells
(A) The SINC accumulates in old mothers. Fluorescence micrographs of Nup85-GFP in _vps4Δpom152_Δ cells with the indicated number of bud scars stained with calcofluor. Bar is 1 µm. (B) Plot of percentage of cells with Nup85-GFP in SINC in reference to bud scar number. Mean +/− s.d. from 3 experiments, n>60. (C) The SINC is retained in mother cells. Images of Nup85-GFP at a mother (M) nucleus and of its four daughter nuclei (D1-D4) in the indicated strains. Fluorescence in each image series is normalized to the fluorescence of the mother cells after the final division to avoid saturating the SINC signal. Bar is 1µm. See Movie S1. (D) NPC clusters of nup120A and nup133A strains are not retained in mothers. Plot of the percentage of divisions that result in the retention of nup clusters in mother. Mean +/− s.d. from 3 experiments. See Movies S2, S3, S4. (E) Nuclear compartmentalization is lost in vps4A mother cells. Plot of the quantification of the relative nuclear accumulation of NLS-GFP between individual mother (M) and daughter (D) cells expressed as a ratio of MNUC:MCYT to DNUC:DCYT. Eleven divisions (mother and daughter) analyzed from 3 experiments. Mean +/− s.d. P values from an unpaired, two-tailed t-test. (F) Daughter cells retain nuclear transport. Fluorescence micrographs of NLS-GFP of a mother (M) and its daughter (D) after division. Fluorescence intensities along indicated lines are plotted. MIP is maximum intensity projection. Bar is 1µm.
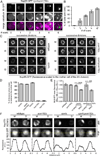
Figure 7. Vps4 destabilizes misassembled nups
(A) An extra pool of Nup85 accumulates in the SINC of _vps4_Δ mothers. Plot of total Nup85-GFP fluorescence in mother (left) and daughter (right) nuclei measured after each of three divisions (normalized to first). Mean +/− s.d. from 3 experiments in which 5 cells (mother and daughter) were measured. (B) Nup85-GFP is stabilized in _vps4_Δ cells when NPC assembly is inhibited. Micrographs of Nup85-GFP at either RT or at 34°C. Bars are 5µm. (C) Nup85-GFP is degraded by the proteasome when NPC assembly is inhibited. Western blot of Nup85-GFP levels (with actin or CPY load controls) from experiments identical to that shown in B. (D) Plot of the quantitation of Nup85-GFP levels in the indicated strains. Mean +/− s.d. from 3 experiments. (E) NUP85 transcript levels decline in _nic96-1vps4_Δ cells. RT-qPCR analysis of NUP85 transcript compared to WT cells. See Figure S5.
Comment in
- ESCRTs take on a job in surveillance.
Odorizzi G. Odorizzi G. Cell. 2014 Oct 9;159(2):240-1. doi: 10.1016/j.cell.2014.09.046. Cell. 2014. PMID: 25303522
Similar articles
- ESCRTs breach the nuclear border.
Webster BM, Lusk CP. Webster BM, et al. Nucleus. 2015;6(3):197-202. doi: 10.1080/19491034.2015.1035844. Epub 2015 May 5. Nucleus. 2015. PMID: 25942571 Free PMC article. - Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing.
Webster BM, Thaller DJ, Jäger J, Ochmann SE, Borah S, Lusk CP. Webster BM, et al. EMBO J. 2016 Nov 15;35(22):2447-2467. doi: 10.15252/embj.201694574. Epub 2016 Oct 12. EMBO J. 2016. PMID: 27733427 Free PMC article. - ESCRTs take on a job in surveillance.
Odorizzi G. Odorizzi G. Cell. 2014 Oct 9;159(2):240-1. doi: 10.1016/j.cell.2014.09.046. Cell. 2014. PMID: 25303522 - The role of VPS4 in ESCRT-III polymer remodeling.
Caillat C, Maity S, Miguet N, Roos WH, Weissenhorn W. Caillat C, et al. Biochem Soc Trans. 2019 Feb 28;47(1):441-448. doi: 10.1042/BST20180026. Epub 2019 Feb 19. Biochem Soc Trans. 2019. PMID: 30783012 Review. - Nuclear pore complex and nucleocytoplasmic transport disruption in neurodegeneration.
Cristi AC, Rapuri S, Coyne AN. Cristi AC, et al. FEBS Lett. 2023 Oct;597(20):2546-2566. doi: 10.1002/1873-3468.14729. Epub 2023 Sep 12. FEBS Lett. 2023. PMID: 37657945 Free PMC article. Review.
Cited by
- Host and Viral Factors Involved in Nuclear Egress of Herpes Simplex Virus 1.
Arii J. Arii J. Viruses. 2021 Apr 25;13(5):754. doi: 10.3390/v13050754. Viruses. 2021. PMID: 33923040 Free PMC article. Review. - Patrolling the nucleus: inner nuclear membrane-associated degradation.
Smoyer CJ, Jaspersen SL. Smoyer CJ, et al. Curr Genet. 2019 Oct;65(5):1099-1106. doi: 10.1007/s00294-019-00971-1. Epub 2019 Apr 24. Curr Genet. 2019. PMID: 31020383 Free PMC article. Review. - Disrupted neuronal trafficking in amyotrophic lateral sclerosis.
Burk K, Pasterkamp RJ. Burk K, et al. Acta Neuropathol. 2019 Jun;137(6):859-877. doi: 10.1007/s00401-019-01964-7. Epub 2019 Feb 5. Acta Neuropathol. 2019. PMID: 30721407 Free PMC article. Review. - Fragile X-Related Protein 1 Regulates Nucleoporin Localization in a Cell Cycle-Dependent Manner.
Agote-Arán A, Lin J, Sumara I. Agote-Arán A, et al. Front Cell Dev Biol. 2021 Dec 16;9:755847. doi: 10.3389/fcell.2021.755847. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34977012 Free PMC article. - Drosophila Wash and the Wash regulatory complex function in nuclear envelope budding.
Verboon JM, Nakamura M, Davidson KA, Decker JR, Nandakumar V, Parkhurst SM. Verboon JM, et al. J Cell Sci. 2020 Jul 8;133(13):jcs243576. doi: 10.1242/jcs.243576. J Cell Sci. 2020. PMID: 32503943 Free PMC article.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. - PubMed
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007223/GM/NIGMS NIH HHS/United States
- R21 HG006742/HG/NHGRI NIH HHS/United States
- R01 GM105672/GM/NIGMS NIH HHS/United States
- 5T32GM007223/GM/NIGMS NIH HHS/United States
- R21HG006742/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous