Biological functions of casein kinase 1 isoforms and putative roles in tumorigenesis - PubMed (original) (raw)
Review
Biological functions of casein kinase 1 isoforms and putative roles in tumorigenesis
Birgit Schittek et al. Mol Cancer. 2014.
Abstract
Isoforms of the casein kinase 1 (CK1) family have been shown to phosphorylate key regulatory molecules involved in cell cycle, transcription and translation, the structure of the cytoskeleton, cell-cell adhesion and receptor-coupled signal transduction. They regulate key signaling pathways known to be critically involved in tumor progression. Recent results point to an altered expression or activity of different CK1 isoforms in tumor cells. This review summarizes the expression and biological function of CK1 family members in normal and malignant cells and the evidence obtained so far about their role in tumorigenesis.
Figures
Figure 1
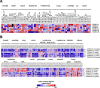
Structure of casein kinase 1 family members. (A) Homology tree of the casein kinase 1 family consisting of six human casein kinase 1 isoforms: alpha, delta, epsilon, gamma1, gamma2 and gamma3. All available and corresponding protein sequences were retrieved from UniProt (see IDs) and the aligned sequences were used to calculate the average distance tree (using the BLOSUM62 algorithm) visualized with jalview. (B) Schematic drawings of the isoforms show conserved regions (yellow color) especially within the kinase domains (dark blue). Variable regions due to transcript variants and alternative splicing are depicted in light blue resulting in variants differing in protein length: CK1α: 337/365 aa; CK1δ: 409/415 aa; CK1ϵ: 416 aa; CK1γ1 393/422 aa; CK1γ2: 415 aa; CK1γ3: 311–455 aa. Phosphorylation sites (red) are occurring predominantly at the C-terminal ends of the delta (Ser331/370/382/383/384/411) and epsilon isoforms (Ser343/354/362/363/389) and are known to be auto-inhibitory. An additional phosphorylation site can be found within the 28 aa insertion of CK1α (Ser156). A nuclear localization signal is located in the long variant of the alpha isoform (aa 160–163). A centrosomal localization signal [7] is located at the C-terminus in the delta (aa 278–364) isoform.
Figure 2
Expression of CK1 isoforms in cancer. (A) Shown are the color-coded RNA expression levels of CK1α, δ, ϵ, γ1, γ2, ϵ3 (CSNK1A1, CSNK1D, CSNK1E, CSNK1G1, CSNK1G2, CSNK1G3) in different tumor types as determined by the CellMiner database for gene expression analysis of 60 cancer cell lines from the NCI60 panel. Z-scores for the six isoforms were calculated and subjected to the Gene-Pattern analysis software for heatmap generation using the Gene-Pattern online tools (
http://genepattern.broadinstitute.org/gp/pages/index.jsf
). Blue depicts low expression levels and red high expression levels. (B) The Ramaswamy Multi-cancer dataset (public available at
http://www.broadinstitute.org/cancer/software/genepattern/datasets/
) was used for analysis of the z-scored expression levels in different tumor tissue samples and again used for heatmap illustration.
Figure 3
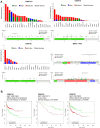
Mutations in CK1 isoforms and cancer patient survival. (A) Mutation analysis of CK1 isoforms and overall survival of tumor patients stratified in CK1 isoform expression classes. 24 different tumor entities were analyzed using the cBioPortal for Cancer Genomics [94, 95] accessing the newest TCGA datasets. The highest mutation frequency for CK1α was detected in clear cell renal cell carcinoma (ccRCC) with approx. 4.8%, for CK1δ in liver cancer with approx. 9.5% and for CK1ϵ with approximately 3.8% (top diagrams). The distribution along the primary structure of the three CK1 isoforms α, δ and ϵ was analyzed in the cBioportal using all available datasets (69 cancer studies containing data of 17584 tumor samples). No hotspot mutation could be identified and only very low mutation frequencies were found (bottom diagrams). In comparison, analysis of the oncogene BRAF and the tumor suppressor TP53 revealed high mutation frequencies and the hot spot mutations at BRAFV600D/E/K/R or TP53R273C/H/L/P/S/Y. (B) Survival analysis of stratified cancer patients into high and low CK1 isoform expression (determined by microarray gene expression analysis) was performed using the PPISURV online tool [96]. The studies showing the highest significant differences in overall survival (OS) between CK1high and CK1low patients are shown as Kaplan Meier curves for the α, δ and ϵ isoforms (CK1 high expression in red, CK1 low expression in green).
Similar articles
- Functions and regulation of the serine/threonine protein kinase CK1 family: moving beyond promiscuity.
Fulcher LJ, Sapkota GP. Fulcher LJ, et al. Biochem J. 2020 Dec 11;477(23):4603-4621. doi: 10.1042/BCJ20200506. Biochem J. 2020. PMID: 33306089 Free PMC article. Review. - CK1 in Developmental Signaling: Hedgehog and Wnt.
Jiang J. Jiang J. Curr Top Dev Biol. 2017;123:303-329. doi: 10.1016/bs.ctdb.2016.09.002. Epub 2016 Oct 26. Curr Top Dev Biol. 2017. PMID: 28236970 Free PMC article. Review. - The casein kinase 1 family: participation in multiple cellular processes in eukaryotes.
Knippschild U, Gocht A, Wolff S, Huber N, Löhler J, Stöter M. Knippschild U, et al. Cell Signal. 2005 Jun;17(6):675-89. doi: 10.1016/j.cellsig.2004.12.011. Epub 2005 Jan 25. Cell Signal. 2005. PMID: 15722192 Review. - The role of the casein kinase 1 (CK1) family in different signaling pathways linked to cancer development.
Knippschild U, Wolff S, Giamas G, Brockschmidt C, Wittau M, Würl PU, Eismann T, Stöter M. Knippschild U, et al. Onkologie. 2005 Oct;28(10):508-14. doi: 10.1159/000087137. Epub 2005 Sep 19. Onkologie. 2005. PMID: 16186692 Review. - Casein kinase 1 and Wnt/β-catenin signaling.
Cruciat CM. Cruciat CM. Curr Opin Cell Biol. 2014 Dec;31:46-55. doi: 10.1016/j.ceb.2014.08.003. Epub 2014 Sep 15. Curr Opin Cell Biol. 2014. PMID: 25200911 Review.
Cited by
- Cytoplasmic CK1ε Protein Expression Is Correlated With Distant Metastasis and Survival in Patients With Melanoma.
Lu JW, Lin SH, Yeh CM, Yeh KT, Huang LR, Chen CY, Lin YM. Lu JW, et al. In Vivo. 2020 Sep-Oct;34(5):2905-2911. doi: 10.21873/invivo.12119. In Vivo. 2020. PMID: 32871831 Free PMC article. - The interplay between HPIP and casein kinase 1α promotes renal cell carcinoma growth and metastasis via activation of mTOR pathway.
Mai H, Xu X, Mei G, Hong T, Huang J, Wang T, Yan Z, Li Y, Liang Y, Li L, Jin S, You W, Ma Y, Chen L, Ye Q. Mai H, et al. Oncogenesis. 2016 Oct 3;5(10):e260. doi: 10.1038/oncsis.2016.44. Oncogenesis. 2016. PMID: 27694835 Free PMC article. - Deep Transcriptomic Analysis Reveals the Dynamic Developmental Progression during Early Development of Channel Catfish (Ictalurus punctatus).
Ma X, Su B, Tian Y, Backenstose NJC, Ye Z, Moss A, Duong TY, Wang X, Dunham RA. Ma X, et al. Int J Mol Sci. 2020 Aug 2;21(15):5535. doi: 10.3390/ijms21155535. Int J Mol Sci. 2020. PMID: 32748829 Free PMC article. - Novel Insights into the Biochemical Mechanism of CK1ε and its Functional Interplay with DDX3X.
Bono B, Franco G, Riva V, Garbelli A, Maga G. Bono B, et al. Int J Mol Sci. 2020 Sep 3;21(17):6449. doi: 10.3390/ijms21176449. Int J Mol Sci. 2020. PMID: 32899434 Free PMC article. - A Mouse Systems Genetics Approach Reveals Common and Uncommon Genetic Modifiers of Hepatic Lysosomal Enzyme Activities and Glycosphingolipids.
Durán A, Priestman DA, Las Heras M, Rebolledo-Jaramillo B, Olguín V, Calderón JF, Zanlungo S, Gutiérrez J, Platt FM, Klein AD. Durán A, et al. Int J Mol Sci. 2023 Mar 3;24(5):4915. doi: 10.3390/ijms24054915. Int J Mol Sci. 2023. PMID: 36902345 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials