δEF1 associates with DNMT1 and maintains DNA methylation of the E-cadherin promoter in breast cancer cells - PubMed (original) (raw)
δEF1 associates with DNMT1 and maintains DNA methylation of the E-cadherin promoter in breast cancer cells
Akihiko Fukagawa et al. Cancer Med. 2015 Jan.
Abstract
Abnormal DNA methylation at the C-5 position of cytosine (5mC) of CpG dinucleotides is a well-known epigenetic feature of cancer. Levels of E-cadherin, which is regularly expressed in epithelial tissues, are frequently reduced in epithelial tumors due to transcriptional repression, sometimes accompanied by hypermethylation of the promoter region. δEF1 family proteins (δEF1/ZEB1 and SIP1/ZEB2), key regulators of the epithelial-mesenchymal transition (EMT), suppress E-cadherin expression at the transcriptional level. We recently showed that levels of mRNAs encoding δEF1 proteins are regulated reciprocally with E-cadherin level in breast cancer cells. Here, we examined the mechanism underlying downregulation of E-cadherin expression in three basal-type breast cancer cells in which the E-cadherin promoter region is hypermethylated (Hs578T) or moderately methylated (BT549 and MDA-MB-231). Regardless of methylation status, treatment with 5-aza-2'-deoxycytidine (5-aza), which inhibits DNA methyltransferases, had no effect on E-cadherin expression. Knockdown of δEF1 and SIP1 resulted in recovery of E-cadherin expression in cells lacking hypermethylation, whereas combined treatment with 5-aza synergistically restored E-cadherin expression, especially when the E-cadherin promoter was hypermethylated. Moreover, δEF1 interacted with DNA methyltransferase 1 (DNMT1) through the Smad-binding domain. Sustained knockdown of δEF1 family proteins reduced the number of 5mC sites in the E-cadherin promoter region, suggesting that these proteins maintain 5mC through interaction with DNMT1 in breast cancer cells. Thus, δEF1 family proteins appear to repress expression of E-cadherin during cancer progression, both directly at the transcriptional level and indirectly at the epigenetic level.
Keywords: Cancer cells; DNA methylation; E-cadherin; EMT; δEF1.
© 2014 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Figures
Figure 1
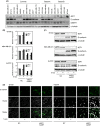
Expression profiles of E-cadherin, _δ_EF1, and DNMT1 in human breast cancer cells. (A) Protein levels of E-cadherin, _δ_EF1, and DNMT1 were determined by immunoblot analysis of whole-cell extracts. _α_-tubulin levels were monitored as a loading control. Molecular subtypes are as reported by Neve et al. . and Charafe-Jauffret et al. . (B, C, and D) BT549, MDA-MB-231, and Hs578T cells were transfected with siRNAs against both _δ_EF1 and SIP1, and then treated with 5 _μ_mol/L of 5-aza-2′-deoxycytidine (5-aza) for 48 h (for BT549 and MDA-MB-231 cells) or 1 _μ_mol/L of 5-aza for 72 h (for Hs587T cells). Cells were then harvested and examined for expression of _δ_EF1, SIP1, and E-cadherin by quantitative RT-PCR (B), immunoblotting (C), or immunocytochemistry (D). NC, negative control.
Figure 2
DNA methylation at the C-5 position (5mC) of cytosine in CpG dinucleotides in human breast cancer cells. (A) Schematic illustration of the promoter region of human E-cadherin (−162 to +37). Numbers in parentheses represent individual CpG sites in the region. E-boxes 1 and 2 have been already reported as binding sites for _δ_EF1 and SIP1–. (B) Bisulfite sequencing was performed using bisulfite-treated templates from MCF7 and T47D cells. White and black circles represent unmethylated and methylated CpG (5mC) sites, respectively. (C) MCF7 and T47D cells were infected with lentiviral vectors encoding FLAG-_δ_EF1. After 48 h, immunoblots were performed on whole-cell extracts. _α_-tubulin levels were monitored as a loading control. (D) Bisulfite sequencing was performed using bisulfite-treated templates from BT549, MDA-MB-231, and Hs578T cells. White and black circles represent unmethylated and methylated CpG (5mC), respectively. (E) The number of 5mC sites was compared among BT549, MDA-MB-231, and Hs578T cells. The Mann–Whitney _U_-test was used for assessing distributional differences of variance across different test samples. *Mann–Whitney _U_-test, P < 0.01. (F) Hs578T cells transfected with siRNAs against both _δ_EF1 and SIP1 were treated with 1 _μ_mol/L 5-aza-2′-deoxycytidine (5-aza) for 72 h. After bisulfite sequencing was performed on 11 clones of Hs578T treated with the indicated combinations, the number of methylated CpG (5mC) sites was counted. Median values are represented by horizontal bars (E and F). NC, negative control.
Figure 3
Interaction of _δ_EF1 with DNMT1. (A–C) HEK293 cells were transiently transfected with the indicated expression plasmids. Twenty-four hours after transfection, cells were harvested, lysed, and subjected to immunoprecipitation (IP) with anti-FLAG antibody, followed by immunoblotting (IB) with anti-Myc antibody. Schematic illustrations depict wild-type (WT), N-terminally truncated mutants (ΔA–ΔD), and C-terminally truncated mutants (ΔE–ΔF) of _δ_EF1 (left panels in B and C). (D) MCF7 and Hs578T cells were harvested and subjected to immunoprecipitation (IP) with anti-DNMT1 antibody or IgG, followed by immunoblotting (IB) with anti-DNMT1 or anti-_δ_EF1 antibodies. _α_-tubulin levels were monitored as a loading control. NZF, N-terminal zinc finger; SBD, Smad-binding domain; HD, homeodomain; CtBP, CtBP-binding domain; CZF, C-terminal zinc finger.
Figure 4
Evaluation of 5mC sites after sustained knockdown of both _δ_EF1 and SIP1 in Hs578T. (A, B, and C) Lentiviral vectors encoding _δ_EF1 and SIP1 shRNAs were used to infect Hs578T cells. Twenty days after infection, the cells were harvested and examined for expression of _δ_EF1/SIP1 and E-cadherin by quantitative RT-PCR (A), immunoblotting (B), or immunofluorescence (C). (D) Schematic illustration of the promoter region of human E-cadherin is shown (top). After bisulfite sequencing was performed, the number of methylated CpG (5mC) sites ay (11)–(17) was counted (right). White and black circles represent unmethylated and methylated CpG (5mC) sites, respectively (left). Median values are represented as horizontal bars (right). NC, negative control.
Similar articles
- Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta.
Shirakihara T, Saitoh M, Miyazono K. Shirakihara T, et al. Mol Biol Cell. 2007 Sep;18(9):3533-44. doi: 10.1091/mbc.e07-03-0249. Epub 2007 Jul 5. Mol Biol Cell. 2007. PMID: 17615296 Free PMC article. - TGF-β drives epithelial-mesenchymal transition through δEF1-mediated downregulation of ESRP.
Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, Fujii H, Yamaguchi A, Miyazawa K, Miyazono K, Saitoh M. Horiguchi K, et al. Oncogene. 2012 Jun 28;31(26):3190-201. doi: 10.1038/onc.2011.493. Epub 2011 Oct 31. Oncogene. 2012. PMID: 22037216 Free PMC article. - The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1.
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. Gregory PA, et al. Nat Cell Biol. 2008 May;10(5):593-601. doi: 10.1038/ncb1722. Epub 2008 Mar 30. Nat Cell Biol. 2008. PMID: 18376396 - SIP1 (Smad interacting protein 1) and deltaEF1 (delta-crystallin enhancer binding factor) are structurally similar transcriptional repressors.
van Grunsven LA, Schellens A, Huylebroeck D, Verschueren K. van Grunsven LA, et al. J Bone Joint Surg Am. 2001;83-A Suppl 1(Pt 1):S40-7. J Bone Joint Surg Am. 2001. PMID: 11263664 Review. - Interactions between E-cadherin and microRNA deregulation in head and neck cancers: the potential interplay.
Wong TS, Gao W, Chan JY. Wong TS, et al. Biomed Res Int. 2014;2014:126038. doi: 10.1155/2014/126038. Epub 2014 Aug 4. Biomed Res Int. 2014. PMID: 25161999 Free PMC article. Review.
Cited by
- Cross-Talk between the TGF-β and Cell Adhesion Signaling Pathways in Cancer.
Liao J, Chen R, Lin B, Deng R, Liang Y, Zeng J, Ma S, Qiu X. Liao J, et al. Int J Med Sci. 2024 May 13;21(7):1307-1320. doi: 10.7150/ijms.96274. eCollection 2024. Int J Med Sci. 2024. PMID: 38818471 Free PMC article. Review. - Plasticity in cell migration modes across development, physiology, and disease.
Pourjafar M, Tiwari VK. Pourjafar M, et al. Front Cell Dev Biol. 2024 Apr 23;12:1363361. doi: 10.3389/fcell.2024.1363361. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38715921 Free PMC article. Review. - MCL1 Inhibition Overcomes the Aggressiveness Features of Triple-Negative Breast Cancer MDA-MB-231 Cells.
Pratelli G, Carlisi D, Di Liberto D, Notaro A, Giuliano M, D'Anneo A, Lauricella M, Emanuele S, Calvaruso G, De Blasio A. Pratelli G, et al. Int J Mol Sci. 2023 Jul 6;24(13):11149. doi: 10.3390/ijms241311149. Int J Mol Sci. 2023. PMID: 37446326 Free PMC article. - The Role of EMT-Related lncRNAs in Ovarian Cancer.
Lampropoulou DI, Papadimitriou M, Papadimitriou C, Filippou D, Kourlaba G, Aravantinos G, Gazouli M. Lampropoulou DI, et al. Int J Mol Sci. 2023 Jun 13;24(12):10079. doi: 10.3390/ijms241210079. Int J Mol Sci. 2023. PMID: 37373222 Free PMC article. Review. - Dissecting the effects of androgen deprivation therapy on cadherin switching in advanced prostate cancer: A molecular perspective.
Varisli L, Tolan V, Cen JH, Vlahopoulos S, Cen O. Varisli L, et al. Oncol Res. 2023 Jan 12;30(3):137-155. doi: 10.32604/or.2022.026074. eCollection 2022. Oncol Res. 2023. PMID: 37305018 Free PMC article. Review.
References
- Ringner M, Staaf J. Jonsson G. Nonfamilial breast cancer subtypes. Methods Mol. Biol. 2013;973:279–295. - PubMed
- Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adélaïde J, Cervera N, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. - PubMed
- Thiery JP, Acloque H, Huang RY. Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous