A new rhesus macaque assembly and annotation for next-generation sequencing analyses - PubMed (original) (raw)
doi: 10.1186/1745-6150-9-20.
Adam S Cornish, Mnirnal D Maudhoo, Robert M Gibbs, Xiongfei Zhang, Sanjit Pandey, Daniel T Meehan, Kristin Wipfler, Steven E Bosinger, Zachary P Johnson, Gregory K Tharp, Guillaume Marçais, Michael Roberts, Betsy Ferguson, Howard S Fox, Todd Treangen, Steven L Salzberg, James A Yorke, Robert B Norgren Jr 1
Affiliations
- PMID: 25319552
- PMCID: PMC4214606
- DOI: 10.1186/1745-6150-9-20
A new rhesus macaque assembly and annotation for next-generation sequencing analyses
Aleksey V Zimin et al. Biol Direct. 2014.
Abstract
Background: The rhesus macaque (Macaca mulatta) is a key species for advancing biomedical research. Like all draft mammalian genomes, the draft rhesus assembly (rheMac2) has gaps, sequencing errors and misassemblies that have prevented automated annotation pipelines from functioning correctly. Another rhesus macaque assembly, CR_1.0, is also available but is substantially more fragmented than rheMac2 with smaller contigs and scaffolds. Annotations for these two assemblies are limited in completeness and accuracy. High quality assembly and annotation files are required for a wide range of studies including expression, genetic and evolutionary analyses.
Results: We report a new de novo assembly of the rhesus macaque genome (MacaM) that incorporates both the original Sanger sequences used to assemble rheMac2 and new Illumina sequences from the same animal. MacaM has a weighted average (N50) contig size of 64 kilobases, more than twice the size of the rheMac2 assembly and almost five times the size of the CR_1.0 assembly. The MacaM chromosome assembly incorporates information from previously unutilized mapping data and preliminary annotation of scaffolds. Independent assessment of the assemblies using Ion Torrent read alignments indicates that MacaM is more complete and accurate than rheMac2 and CR_1.0. We assembled messenger RNA sequences from several rhesus tissues into transcripts which allowed us to identify a total of 11,712 complete proteins representing 9,524 distinct genes. Using a combination of our assembled rhesus macaque transcripts and human transcripts, we annotated 18,757 transcripts and 16,050 genes with complete coding sequences in the MacaM assembly. Further, we demonstrate that the new annotations provide greatly improved accuracy as compared to the current annotations of rheMac2. Finally, we show that the MacaM genome provides an accurate resource for alignment of reads produced by RNA sequence expression studies.
Conclusions: The MacaM assembly and annotation files provide a substantially more complete and accurate representation of the rhesus macaque genome than rheMac2 or CR_1.0 and will serve as an important resource for investigators conducting next-generation sequencing studies with nonhuman primates.
Reviewers: This article was reviewed by Dr. Lutz Walter, Dr. Soojin Yi and Dr. Kateryna Makova.
Figures
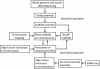
Figure 1
Flowchart illustrating procedures for assembly and annotation of the MacaM rhesus macaque genome.
Figure 2
Correction of rheMac2 SHE gene misassembly in MacaM. A. rheMac2 genome. Exons 1, 2, 4, 5 and 6 of the Src homology 2 domain containing E. (SHE) gene are contained within scaffold NW_001108937.1. Exon 3 of this gene was assigned to scaffold NW_001218118.1. Scaffold NW_001108937.1 was correctly assigned to chromosome 1. However, scaffold NW_001218118.1 was mistakenly assigned to chromosome X. This resulted in an annotation of the rhesus SHE gene with missing sequence (corresponding to exon 3). Additional details on the misassembly of this gene in rheMac2 can be found in [3]. B. MacaM genome. All 6 exons of the SHE gene were found on scaffold 2317188291 of the MacaM assembly.
Figure 3
Alignment of rhesus macaque SHE proteins from different annotations with human protein. Human SHE protein accession: NP_001010846.1. MacaM: Protein derived from the MacaM rhesus macaque genome. rheMac2_N: Protein obtained from the NCBI annotation of rheMac2, accession. rheMac2_E: Protein obtained from the Ensembl annotation of rheMac2, accession ENSMMUT00000032345. CR_1.0: Protein obtained from the Chinese rhesus macaque genome produced by BGI [8]. Yellow highlighting indicates identical sequence in human and alternative rhesus macaque annotations with the exception of sequences that are only shared in rheMac2_E and CR_1.0 which are indicated by green highlighting. Exon boundaries are indicated by line separating amino acids.
Figure 4
mRNA expression validation. We sequenced RNA from 60 rhesus macaque PBMC samples of differing ranks using Illumina paired end sequencing. After filtering, we mapped reads to either the MacaM (green symbols) or rheMac2 (blue symbols) assemblies using the STAR algorithm; we used CUFFLINKS to assign transcripts and determine differentially expressed genes (DEGs). (A) Number of uniquely mapping reads in individual RNA samples mapped using the MacaM and rheMac2 assemblies. Individual samples mapped by either assembly are joined by lines. (B) Percentage of total filtered reads that uniquely mapped to each assembly. (C) Number of DEGs that were identified using CUFFDIFF2.1 for dominant animals at two time points using the MacaM and rheMac2 genomes.
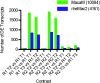
Figure 5
Number of DEGs which were identified in an experiment analyzing social anxiety in rhesus macaques. CUFFDIFF2.1 was used to identify DEGs with two Ranks (R1 = dominant; R2 = subordinate) and three time points (T1 = baseline; T2 = T1 + 20 minutes; T3 = T1 + 260 minutes). Human intruder intervention occurred immediately before T2, after T1.
Similar articles
- Exome screening to identify loss-of-function mutations in the rhesus macaque for development of preclinical models of human disease.
Cornish AS, Gibbs RM, Norgren RB Jr. Cornish AS, et al. BMC Genomics. 2016 Mar 2;17:170. doi: 10.1186/s12864-016-2509-5. BMC Genomics. 2016. PMID: 26935327 Free PMC article. - Advantages of an Improved Rhesus Macaque Genome for Evolutionary Analyses.
Gradnigo JS, Majumdar A, Norgren RB Jr, Moriyama EN. Gradnigo JS, et al. PLoS One. 2016 Dec 2;11(12):e0167376. doi: 10.1371/journal.pone.0167376. eCollection 2016. PLoS One. 2016. PMID: 27911958 Free PMC article. - Limitations of the rhesus macaque draft genome assembly and annotation.
Zhang X, Goodsell J, Norgren RB Jr. Zhang X, et al. BMC Genomics. 2012 May 30;13:206. doi: 10.1186/1471-2164-13-206. BMC Genomics. 2012. PMID: 22646658 Free PMC article. - Improving genome assemblies and annotations for nonhuman primates.
Norgren RB Jr. Norgren RB Jr. ILAR J. 2013;54(2):144-53. doi: 10.1093/ilar/ilt037. ILAR J. 2013. PMID: 24174438 Free PMC article. Review. - Scalable and versatile container-based pipelines for de novo genome assembly and bacterial annotation.
de Almeida FM, de Campos TA, Pappas GJ Jr. de Almeida FM, et al. F1000Res. 2023 Sep 25;12:1205. doi: 10.12688/f1000research.139488.1. eCollection 2023. F1000Res. 2023. PMID: 37970066 Free PMC article. Review.
Cited by
- Characterization of the plasma proteome of nonhuman primates during Ebola virus disease or melioidosis: a host response comparison.
Ward MD, Brueggemann EE, Kenny T, Reitstetter RE, Mahone CR, Trevino S, Wetzel K, Donnelly GC, Retterer C, Norgren RB Jr, Panchal RG, Warren TK, Bavari S, Cazares LH. Ward MD, et al. Clin Proteomics. 2019 Feb 7;16:7. doi: 10.1186/s12014-019-9227-3. eCollection 2019. Clin Proteomics. 2019. PMID: 30774579 Free PMC article. - Genetic Architecture of Human Obesity Traits in the Rhesus Macaque.
Raboin MJ, Letaw J, Mitchell AD, Toffey D, McKelvey J, Roberts CT Jr, Curran JE, Vinson A. Raboin MJ, et al. Obesity (Silver Spring). 2019 Mar;27(3):479-488. doi: 10.1002/oby.22392. Epub 2019 Feb 11. Obesity (Silver Spring). 2019. PMID: 30741480 Free PMC article. - Changes in gene expression following long-term in vitro exposure of Macaca mulatta trophoblast stem cells to biologically relevant levels of endocrine disruptors.
Midic U, Goheen B, Vincent KA, VandeVoort CA, Latham KE. Midic U, et al. Reprod Toxicol. 2018 Apr;77:154-165. doi: 10.1016/j.reprotox.2018.02.012. Epub 2018 Mar 2. Reprod Toxicol. 2018. PMID: 29505797 Free PMC article. - Caloric Restriction Engages Hepatic RNA Processing Mechanisms in Rhesus Monkeys.
Rhoads TW, Burhans MS, Chen VB, Hutchins PD, Rush MJP, Clark JP, Stark JL, McIlwain SJ, Eghbalnia HR, Pavelec DM, Ong IM, Denu JM, Markley JL, Coon JJ, Colman RJ, Anderson RM. Rhoads TW, et al. Cell Metab. 2018 Mar 6;27(3):677-688.e5. doi: 10.1016/j.cmet.2018.01.014. Cell Metab. 2018. PMID: 29514073 Free PMC article. - Caloric restriction improves health and survival of rhesus monkeys.
Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, Roth GS, Ingram DK, Weindruch R, de Cabo R, Anderson RM. Mattison JA, et al. Nat Commun. 2017 Jan 17;8:14063. doi: 10.1038/ncomms14063. Nat Commun. 2017. PMID: 28094793 Free PMC article.
References
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y. et al.Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases