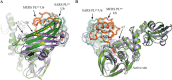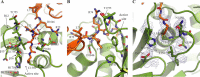Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression - PubMed (original) (raw)
Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression
Ben A Bailey-Elkin et al. J Biol Chem. 2014.
Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) is a newly emerging human pathogen that was first isolated in 2012. MERS-CoV replication depends in part on a virus-encoded papain-like protease (PL(pro)) that cleaves the viral replicase polyproteins at three sites releasing non-structural protein 1 (nsp1), nsp2, and nsp3. In addition to this replicative function, MERS-CoV PL(pro) was recently shown to be a deubiquitinating enzyme (DUB) and to possess deISGylating activity, as previously reported for other coronaviral PL(pro) domains, including that of severe acute respiratory syndrome coronavirus. These activities have been suggested to suppress host antiviral responses during infection. To understand the molecular basis for ubiquitin (Ub) recognition and deconjugation by MERS-CoV PL(pro), we determined its crystal structure in complex with Ub. Guided by this structure, mutations were introduced into PL(pro) to specifically disrupt Ub binding without affecting viral polyprotein cleavage, as determined using an in trans nsp3↓4 cleavage assay. Having developed a strategy to selectively disable PL(pro) DUB activity, we were able to specifically examine the effects of this activity on the innate immune response. Whereas the wild-type PL(pro) domain was found to suppress IFN-β promoter activation, PL(pro) variants specifically lacking DUB activity were no longer able to do so. These findings directly implicate the DUB function of PL(pro), and not its proteolytic activity per se, in the inhibition of IFN-β promoter activity. The ability to decouple the DUB activity of PL(pro) from its role in viral polyprotein processing now provides an approach to further dissect the role(s) of PL(pro) as a viral DUB during MERS-CoV infection.
Keywords: Cysteine Protease; Deubiquitylation (Deubiquitination); Innate Immunity; Middle East Respiratory Syndrome Coronavirus; PLpro; Structural Biology; Viral Immunology; X-ray Crystallography.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
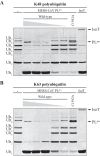
FIGURE 1.
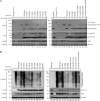
In vitro cleavage of Lys48- and Lys63-linked poly-Ub chains by recombinant MERS-CoV PLpro. Purified recombinant MERS-CoV PLpro was incubated with 2.5 μg of Lys48-linked (A) or Lys63-linked (B) poly-Ub chains of different length in each reaction for 2 h at 37 °C in a final volume of 10 μl. A range of 2-fold dilutions starting at 2 μ
m
MERS-CoV wild-type PLpro per reaction was used. Activity of the PLpro active site mutant (C1592A) was assessed at a concentration of 2 μ
m
. Isopeptidase T (IsoT; 0.5 μg/reaction) served as a positive control (69).
FIGURE 2.
MERS-CoV PLpro and PLpro·Ub structures. A, structure of the MERS-CoV PLprodomain (2.15 Å resolution). The palm, thumb, fingers, and N-terminal ubiquitin-like (Ubl) domains are indicated by_colored panels_, and arrows_indicate the active site and C4 zinc ribbon motif. The active site residues are depicted as sticks. B, structure of the MERS-CoV PLpro bound to Ub (2.8 Å resolution). PLpro is shown in_green, and the covalently bound Ub molecule is orange and shown as tubes. Active site residues are shown as sticks with Gly75 and the 3CN linker of Ub covalently linked to Cys1592 of PLpro. C, superposition showing a ∼6.8-Å movement of the zinc ribbon motif between the open (yellow) and closed (green) PLpro·Ub structures and a previously reported PLpro structure (gray) (Protein Data Bank entry 4P16 (49)). Our PLprostructure is not shown because it is highly similar to the open PLpro·Ub structure. Movement of the zinc ribbon motif was determined by measuring the distance between the zinc atom of the respective structures. Superpositions were performed in Coot (45). Ub was removed from the closed and open PLpro·Ub structures for clarity. Figures were created using PyMOL (70).
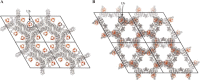
FIGURE 3.
Crystal packing arrangement of the open and closed MERS-CoV PLpro·Ub structures. The contents of four unit cells are shown, with PLpro and Ub depicted in gray and_orange_, respectively. A, the open PLpro·Ub structure crystallized in space group P63, where Ub was found to face the solvent, uninvolved in crystal contacts. B, the closed PLpro·Ub structure crystallized in space group P6522, where Ub no longer faces the solvent, and is involved in crystal contacts. Images were created using PyMOL (70).
FIGURE 4.
Structural comparison of the SARS-CoV PLpro·Ub and MERS-CoV PLpro·Ub complexes. A, superposition of the closed MERS-CoV PLpro·Ub complex (green) and the SARS-CoV PLpro·Ub complex (purple; Protein Data Bank entry 4M0W) using SSM superpose in Coot (45) (bound Ub molecules were ignored during the superposition). The Ub molecules bound to the MERS-CoV PLpro domain and SARS-CoV PLpro domain are depicted as tubes in_orange_ and pale cyan, respectively. The ∼26° shift in the fingers domain between the two respective structures is indicated.B, alternate orientation of the SARS-CoV PLpro·Ub and MERS-CoV PLpro·Ub superpositions highlighting the difference in Ub binding. In the MERS-CoV PLpro·Ub complex, Ub is found shifted toward helix α7 compared with the SARS PLpro·Ub complex. Helix α7 of MERS-CoV PLpro is indicated with an_arrow_. Images were created using PyMOL (70).
FIGURE 5.
Active site of MERS-CoV PLpro and interactions with the C-terminal RLRGG motif of Ub. Interactions between open PLpro (green) and the C-terminal RLRGG motif of Ub (orange) are depicted in A_and B. A, the main-chain amide of the 3CN linker, which mimics Gly76 of Ub, forms a hydrogen bond with the main chain carbonyl of PLpro residue Gly1758. The main-chain amide of Gly75 of Ub forms a hydrogen bond with the carbonyl group of PLpro Asp1645, and a hydrogen bonding interaction occurs between the main-chain carbonyl of Arg74of Ub and the main-chain amide of Gly1758 of PLpro. The side-chain η-amino group of Ub residue Arg74 is hydrogen-bonded to the main-chain carbonyl group of PLpro Thr1755. Hydrogen bonds also occur between the side-chain ϵ- and η-amino groups of Ub Arg72 and the carboxylate of PLproAsp1645 as well as between the main-chain amide of Ub residue Leu73 and side chain PLpro residue Asp1646. The BL2 loop between strands β15 and β16 is indicated with an arrow. B, alternate orientation of the PLpro active site showing a hydrogen bonding interaction between the Ub Leu73 main-chain amide group and the side-chain carboxylate of PLpro residue Asp1646. The side chain of Ub residue Leu63also undergoes hydrophobic interactions with PLpro residues Phe1750 and Pro1731. C, the MERS-CoV PLproCys1592-His1759-Asp1774 catalytic triad residues are shown as well as residues Asn1590 and Asn1591, which together with Cys1592 form the oxyanion hole via their backbone amide groups. The covalent 3CN molecule is shown linking the C terminus of Ub to the active site Cys1592 of PLpro. The active site Leu1587 residue, which is not involved in oxyanion hole formation, is also shown. The electron density is a maximum likelihood weighted 2_Fo −Fc map contoured at 1.0σ. Images were created using PyMOL (70).
FIGURE 6.
Structure-guided mutagenesis of PLpro residues involved in Ub recognition. A, surface representation of the closed MERS-CoV PLpro·Ub complex. PLpro is shown in green, and Ub is shown in transparent orange. Those residues that were mutated in order to disrupt Ub binding are colored magenta and are indicated with arrows. Colored boxes refer to close-up views of the PLpro·Ub interactions and are shown to the right. B, hydrophobic interaction is shown between Val1691 of PLpro and Ile44 of Ub. C, Thr1653 of PLpro is shown hydrogen-bonded to Gln49 and Glu51 of Ub, and Arg1649 of PLprois shown interacting with Arg72 of Ub. D, hydrogen-bonding interactions are shown between Gln1708 of PLpro and Gln62 of Ub, and a hydrophobic interaction is shown between Val1706 of PLpro and Phe4 of Ub. Asn1673 and Val1674 of PLpro, which do not interact with Ub, are also displayed. Images were created using PyMOL (70).
FIGURE 7.
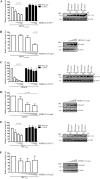
Effect of PLpro mutations on in _trans_cleavage of nsp3↓4 and on DUB activity. A, HEK293T cells were co-transfected with plasmids encoding HA-nsp3C-4-V5 (which does not contain PLpro), PLpro-V5 (wild type and mutants), and GFP (as a transfection control). As a control, plasmid encoding HA-nsp3-4-V5, which includes the PLpro domain, was transfected (lanes 1 and 15), and cleavage resulted in the generation of full-length HA-tagged nsp3 and V5-tagged nsp4. Cells were lysed 18 h post-transfection, and expressed proteins were analyzed by Western blotting. Proteolytic cleavage was measured from the generation of N-terminal HA-tagged nsp3C and C-terminal V5-tagged nsp4. B, HEK293T cells were transfected with a combination of plasmids encoding FLAG-Ub, PLpro-V5 (wild-type and mutants), and GFP (as a transfection control). Cells were lysed 18 h post-transfection, and expressed proteins were analyzed by Western blotting to visualize the deconjugation of FLAG-tagged Ub from a wide range of cellular proteins by MERS-CoV PLpro wild-type and mutants.
FIGURE 8.
MERS-CoV PLpro inhibits RIG-I- and MAVS-induced IFN-β promoter activity. HEK293T cells were transfected with a combination of plasmids expressing a firefly luciferase reporter gene under control of the IFN-β promoter,Renilla luciferase; innate immune response inducers RIG-I(2CARD), MAVS, or IRF3(5D); and increasing amounts of MERS-CoV PLpro wild-type, active site mutant C1592A (A, C, and_E_), or full-length MERS-CoV nsp3 (B, D, and F). Upon induction of the innate immune response with RIG-I(2CARD) and IRF3(5D), cells were transfected with the PLpro (0, 150, 350, or 500 ng) or nsp3 (0, 350, 500, of 1000 ng) constructs. Upon induction with MAVS, cells were transfected with the PLpro (0, 50, 75, 100 or 150 ng) or nsp3 (0, 150, 350 or 500 ng) constructs. At 16 h post-transfection, cells were lysed, and luciferase activity was measured. All experiments were repeated independently at least four times. Significance was evaluated using an unpaired two-tailed Student's _t_test; p values of <0.05 were considered significant. Bars, mean; error bars, S.D. Western blotting was used to verify expression of MERS-CoV PLpro and nsp3.
FIGURE 9.
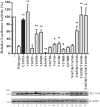
DUB activity is required for IFN-β promoter antagonism by MERS-CoV PLpro. HEK293T cells were transfected with plasmids encoding a firefly luciferase reporter gene under control of the IFN-β promoter, Renilla luciferase, innate immune response inducer MAVS (25 ng), and MERS-CoV PLpro wild type and mutants (75 ng). At 16 h post-transfection, cells were lysed, and luciferase activity was measured. All experiments were repeated independently at least four times. Significance relative to wild type was evaluated using an unpaired two-tailed Student's t test; significant values were indicated as follows: *, p < 0.05; **, p < 0.01. Bars, mean;error bars, S.D. Western blotting was used to verify expression of MERS-CoV PLpro.
Similar articles
- Structural and mutational analysis of the interaction between the Middle-East respiratory syndrome coronavirus (MERS-CoV) papain-like protease and human ubiquitin.
Lei J, Hilgenfeld R. Lei J, et al. Virol Sin. 2016 Aug;31(4):288-99. doi: 10.1007/s12250-016-3742-4. Epub 2016 May 30. Virol Sin. 2016. PMID: 27245450 Free PMC article. - Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease.
Yang X, Chen X, Bian G, Tu J, Xing Y, Wang Y, Chen Z. Yang X, et al. J Gen Virol. 2014 Mar;95(Pt 3):614-626. doi: 10.1099/vir.0.059014-0. Epub 2013 Dec 20. J Gen Virol. 2014. PMID: 24362959 - The emerging SARS-CoV-2 papain-like protease: Its relationship with recent coronavirus epidemics.
Kandeel M, Kitade Y, Fayez M, Venugopala KN, Ibrahim A. Kandeel M, et al. J Med Virol. 2021 Mar;93(3):1581-1588. doi: 10.1002/jmv.26497. Epub 2020 Sep 28. J Med Virol. 2021. PMID: 32902889 - The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds.
Báez-Santos YM, St John SE, Mesecar AD. Báez-Santos YM, et al. Antiviral Res. 2015 Mar;115:21-38. doi: 10.1016/j.antiviral.2014.12.015. Epub 2014 Dec 29. Antiviral Res. 2015. PMID: 25554382 Free PMC article. Review. - Spatial and temporal roles of SARS-CoV PLpro -A snapshot.
Yan S, Wu G. Yan S, et al. FASEB J. 2021 Jan;35(1):e21197. doi: 10.1096/fj.202002271. FASEB J. 2021. PMID: 33368679 Free PMC article. Review.
Cited by
- Drug Development and Medicinal Chemistry Efforts toward SARS-Coronavirus and Covid-19 Therapeutics.
Ghosh AK, Brindisi M, Shahabi D, Chapman ME, Mesecar AD. Ghosh AK, et al. ChemMedChem. 2020 Jun 4;15(11):907-932. doi: 10.1002/cmdc.202000223. Epub 2020 May 7. ChemMedChem. 2020. PMID: 32324951 Free PMC article. Review. - Competing endogenous RNA network profiling reveals novel host dependency factors required for MERS-CoV propagation.
Zhang X, Chu H, Wen L, Shuai H, Yang D, Wang Y, Hou Y, Zhu Z, Yuan S, Yin F, Chan JF, Yuen KY. Zhang X, et al. Emerg Microbes Infect. 2020 Dec;9(1):733-746. doi: 10.1080/22221751.2020.1738277. Emerg Microbes Infect. 2020. PMID: 32223537 Free PMC article. - Middle East Respiratory Syndrome Virus Pathogenesis.
Singh SK. Singh SK. Semin Respir Crit Care Med. 2016 Aug;37(4):572-7. doi: 10.1055/s-0036-1584796. Epub 2016 Aug 3. Semin Respir Crit Care Med. 2016. PMID: 27486737 Free PMC article. Review. - Demonstrating the importance of porcine reproductive and respiratory syndrome virus papain-like protease 2 deubiquitinating activity in viral replication by structure-guided mutagenesis.
Bailey-Elkin BA, Knaap RCM, De Silva A, Boekhoud IM, Mous S, van Vught N, Khajehpour M, van den Born E, Kikkert M, Mark BL. Bailey-Elkin BA, et al. PLoS Pathog. 2023 Dec 14;19(12):e1011872. doi: 10.1371/journal.ppat.1011872. eCollection 2023 Dec. PLoS Pathog. 2023. PMID: 38096325 Free PMC article. - Covid-19: Perspectives on Innate Immune Evasion.
Taefehshokr N, Taefehshokr S, Hemmat N, Heit B. Taefehshokr N, et al. Front Immunol. 2020 Sep 30;11:580641. doi: 10.3389/fimmu.2020.580641. eCollection 2020. Front Immunol. 2020. PMID: 33101306 Free PMC article. Review.
References
- Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D. M. E., Fouchier R. A. M. (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 - PubMed
- World Health Organization (2014) Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Update (United Arab Emirates). World Health Organization, Geneva
- World Health Organization (2003) Summary of Probable SARS Cases with Onset of Illness from 1 November 2002 to 31 July 2003. World Health Organization, Geneva
- Azhar E. I., El-Kafrawy S. A., Farraj S. A., Hassan A. M., Al-Saeed M. S., Hashem A. M., Madani T. A. (2014) Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 370, 2499–2505 - PubMed
- Reusken C. B., Farag E. A., Jonges M., Godeke G. J., El-Sayed A. M., Pas S. D., Raj V. S., Mohran K. J., Moussa H. A., Ghobashy H., Alhajri F., Ibrahim A. K., Bosch B. J., Pasha S. K., Al-Romaihi H. E., Al-Thani M., Al-Marri S. A., AlHajri M. M., Haagmans B. L., Koopmans M. P. (2014) Middle East respiratory syndrome coronavirus (MERS-CoV) RNA and neutralising antibodies in milk collected according to local customs from dromedary camels, Qatar, April 2014. Euro Surveill. 19, 20829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous