Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9 - PubMed (original) (raw)
Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9
Alberto Stolfi et al. Development. 2014 Nov.
Abstract
The CRISPR/Cas9 system has ushered in a new era of targeted genetic manipulations. Here, we report the use of CRISPR/Cas9 to induce double-stranded breaks in the genome of the sea squirt Ciona intestinalis. We use electroporation to deliver CRISPR/Cas9 components for tissue-specific disruption of the Ebf (Collier/Olf/EBF) gene in hundreds of synchronized Ciona embryos. Phenotyping of transfected embryos in the 'F0' generation revealed that endogenous Ebf function is required for specification of Islet-expressing motor ganglion neurons and atrial siphon muscles. We demonstrate that CRISPR/Cas9 is sufficiently effective and specific to generate large numbers of embryos carrying mutations in a targeted gene of interest, which should allow for rapid screening of gene function in Ciona.
Keywords: Ascidians; CRISPR/Cas9; Ciona; Ebf; Genome editing; Islet.
© 2014. Published by The Company of Biologists Ltd.
Figures
Fig. 1.
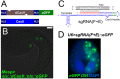
CRISPR/Cas9 components in Ciona intestinalis. (A) Top: nls::dCas9::nls::eGFP protein used to assay nuclear localization in Ciona. dCas9 is Cas9 with two point mutations that render it catalytically dead. Bottom: nls::Cas9::nls used for targeted mutagenesis. (B) Tail bud-stage embryo electroporated with Mesp>nls::dCas9::nls::eGFP, confirming proper nuclear localization in B7.5 lineage cells. Scale bar: 25 μm. (C) Ebf.774 sgRNA (F+E version). The protospacer (red) is paired with its target in Ebf. ‘F’ modification is in orange, ‘E’ modification is in green. (D) In situ hybridization using eGFP probe in gastrula-stage embryo electroporated, with U6>sgRNA(F+E)::eGFP in right hemisphere, indicating successful transcription of the sgRNA in 100% of embryos (_n_=100).
Fig. 2.
CRISPR/Cas9-mediated mutagenesis of Ebf. (A) C. intestinalis Ebf gene, showing exons (solid boxes) and introns (scale bar: 1 kb). Exons are colored according to domains: green, N terminus+DNA-binding domain (DBD); orange, IPT; magenta, atypical HLH; brown, transactivation domain; gray, untranslated regions. Promoters (proximal and distal) are indicated by elbows. Alternative splicing is indicated by dotted line, giving rise to Ebf transcript variants shown below. Both contain the conserved zinc-coordinating motif of the DBD. (B) Alignment of wild-type and mutant Ebf alleles cloned from pooled embryos electroporated with 10 μg EF1α>nls::Cas9::nls, 10 μg U6>Ebf.774 and 10 μg U6>Ebf.813. Three out of seven clones had a mutation (two unique mutations). (C) Mutant Ebf alleles cloned from MACS-sorted hCD4+ cells dissociated from embryos electroporated with 10 μg EF1α>hCD4::mCherry, 25 μg EF1α>nls::Cas9::nls and 75 μg U6>Ebf.774 (see ‘Cleavage assay of genomic DNA from MACS-enriched transfected cells’ section for details). Six out of 13 clones had a mutation (all unique). Target sequences indicated in blue. Indels and substitutions are indicated in red.
Fig. 3.
Genomic cleavage assays. (A) Cleavage assay of Ebf exon 9 amplicon from pooled embryos electroporated with EF1α>nls::Cas9::nls+U6>Ebf.774 or EF1α>nls::Cas9::nls alone. Cleavage efficiency was calculated at 31.5%. Cleavage of Ebf exon 9 amplicon from control (_Cas9-_alone) embryos was not detected. Cleavage of kit control amplicon (1:1 mix of wild-type and mutant sequences) was 43.7%. (B) Assay of amplicon from MACS-sorted cells from embryos electroporated with 10 μg EF1α>nls::Cas9::nls, 10 μg U6>Ebf.774 and 10 μg EF1α>hCD4::mCherry (for MACS selection). Cleavage efficiency in sorted hCD4+ cells was 27.1%, versus 2.4% in hCD4− flow-through and 13.7% in unsorted cells from the same pool of dissociated embryos. (C) Assay of amplicon from MACS-sorted cells from embryos electroporated with 10 μg EF1α>hCD4::mCherry, 25 μg EF1α>nls::Cas9::nls and 75 μg U6>Ebf.774. Cleavage efficiency in sorted hCD4+ cells was 66.2% and 45.1% in unsorted cells from the same embryo pool (see
supplementary material Fig. S6
). (D) Assay of pooled embryos electroporated with EF1α>nls::Cas9::nls+U6>Ebf.774, collected at 4, 8 and 12 hpf. Cleavage bands are visible at 8 hpf, but not at 4 hpf. Efficiency does not increase substantially from 8 to 12 hpf.
Fig. 4.
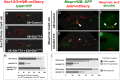
Phenotypic assays for tissue-specific loss of Ebf in F0 embryos. (A) Larva (17.5 hpf, 22°C) electroporated with 35 μg Sox1/2/3>nls::Cas9::nls and 25 μg U6>Control sgRNA, showing normal expression of Isl>YFP reporter in A10.57 MN (observed in 68% of larvae, _n_=100). (B) CRISPR/Cas9-targeted mutagenesis of Ebf exon 9 (35 μg Sox1/2/3>nls::Cas9::nls, 25 μg U6>Ebf.774) results in only 23% of larvae showing MN-specific Isl reporter expression (_n_=100). (C) Lost Isl expression can be rescued by a CRISPR/Cas9-resistant form of Ebf (45 μg Ebf>Ebfm774), as Isl>YFP is now seen in 83% of larvae (_n_=100). (D) Larva (26 hpf, 18°C) electroporated with 50 μg Mesp>nls::Cas9::nls and 25 μg U6>Control, showing normal ASMP migration (seen in 96% of larvae, _n_=100) and _Isl_>mCherry reporter expression in ASMPs (filled arrowhead; seen in 53% of larvae, _n_=100). (E) Upon electroporation with 50 μg Mesp>nls::Cas9::nls and 25 μg U6>Ebf.774, ASMP migration is observed in only 22% of larvae (_n_=100) and _Isl_>mCherry expression in ASMPs is only seen in 3% of larvae (_n_=100; empty arrowhead). Asterisk indicates _Isl_>mCherry expression in A10.57 neuron, confirming that disruption of Ebf was B7.5 lineage specific. (F) Control larvae (22 hpf, 18°C) show normal expression of Mrf (assayed by in situ mRNA hybridization) in ASMPs (filled arrowheads; observed in 79% of larvae, _n_=100). (G) Mrf expression in ASMPs is downregulated upon CRISPR/Cas9 targeting of Ebf (50 μg Mesp>nls::Cas9::nls+25 μg U6>Ebf.774), as Mrf expression is seen in only 38% of larvae (_n_=100; empty arrowheads). (H,I) Histograms indicating the fraction of electroporated embryos displaying the phenotypes represented in A-G.
Similar articles
- CRISPR Knockouts in Ciona Embryos.
Gandhi S, Razy-Krajka F, Christiaen L, Stolfi A. Gandhi S, et al. Adv Exp Med Biol. 2018;1029:141-152. doi: 10.1007/978-981-10-7545-2_13. Adv Exp Med Biol. 2018. PMID: 29542087 Free PMC article. Review. - Optimizing CRISPR/Cas9 approaches in the polymorphic tunicate Ciona intestinalis.
Pennati A, Jakobi M, Zeng F, Ciampa L, Rothbächer U. Pennati A, et al. Dev Biol. 2024 Jun;510:31-39. doi: 10.1016/j.ydbio.2024.03.003. Epub 2024 Mar 14. Dev Biol. 2024. PMID: 38490564 - Evaluation and rational design of guide RNAs for efficient CRISPR/Cas9-mediated mutagenesis in Ciona.
Gandhi S, Haeussler M, Razy-Krajka F, Christiaen L, Stolfi A. Gandhi S, et al. Dev Biol. 2017 May 1;425(1):8-20. doi: 10.1016/j.ydbio.2017.03.003. Epub 2017 Mar 22. Dev Biol. 2017. PMID: 28341547 Free PMC article. - CRISPR/Cas9-mediated gene knockout in the ascidian Ciona intestinalis.
Sasaki H, Yoshida K, Hozumi A, Sasakura Y. Sasaki H, et al. Dev Growth Differ. 2014 Sep;56(7):499-510. doi: 10.1111/dgd.12149. Epub 2014 Sep 12. Dev Growth Differ. 2014. PMID: 25212715 Free PMC article. - Advances in therapeutic CRISPR/Cas9 genome editing.
Savić N, Schwank G. Savić N, et al. Transl Res. 2016 Feb;168:15-21. doi: 10.1016/j.trsl.2015.09.008. Epub 2015 Sep 26. Transl Res. 2016. PMID: 26470680 Review.
Cited by
- MiR-92 Family Members Form a Cluster Required for Notochord Tubulogenesis in Urochordate Ciona savignyi.
Yang L, Zhang X, Liu C, Zhang J, Dong B. Yang L, et al. Genes (Basel). 2021 Mar 12;12(3):406. doi: 10.3390/genes12030406. Genes (Basel). 2021. PMID: 33809016 Free PMC article. - Tunicates: exploring the sea shores and roaming the open ocean. A tribute to Thomas Huxley.
Lemaire P, Piette J. Lemaire P, et al. Open Biol. 2015 Jun;5(6):150053. doi: 10.1098/rsob.150053. Open Biol. 2015. PMID: 26085517 Free PMC article. Review. - Ring Finger 149-Related Is an FGF/MAPK-Independent Regulator of Pharyngeal Muscle Fate Specification.
Vitrinel B, Vogel C, Christiaen L. Vitrinel B, et al. Int J Mol Sci. 2023 May 16;24(10):8865. doi: 10.3390/ijms24108865. Int J Mol Sci. 2023. PMID: 37240211 Free PMC article. - Embryo Microinjection and Electroporation in the Chordate Ciona intestinalis.
Kari W, Zeng F, Zitzelsberger L, Will J, Rothbächer U. Kari W, et al. J Vis Exp. 2016 Oct 16;(116):54313. doi: 10.3791/54313. J Vis Exp. 2016. PMID: 27805579 Free PMC article. - Transgenic Techniques for Investigating Cell Biology During Development.
Cota CD. Cota CD. Adv Exp Med Biol. 2018;1029:153-164. doi: 10.1007/978-981-10-7545-2_14. Adv Exp Med Biol. 2018. PMID: 29542088 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials