Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation - PubMed (original) (raw)
Comparative Study
Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation
Konstantinos Billis et al. PLoS One. 2014.
Abstract
Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803 are model cyanobacteria from which the metabolism and adaptive responses of other cyanobacteria are inferred. Using stranded and 5' enriched libraries, we measured the gene expression response of cells transferred from reference conditions to stress conditions of decreased inorganic carbon, increased salinity, increased pH, and decreased illumination at 1-h and 24-h after transfer. We found that the specific responses of the two strains were by no means identical. Transcriptome profiles allowed us to improve the structural annotation of the genome i.e. identify possible missed genes (including anti-sense), alter gene coordinates and determine transcriptional units (operons). Finally, we predicted associations between proteins of unknown function and biochemical pathways by revealing proteins of known functions that are co-regulated with the unknowns. Future studies of these model organisms will benefit from the cataloging of their responses to environmentally relevant stresses, and improvements in their genome annotations found here.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
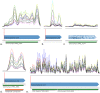
Figure 1. Regions of the genome combined with annotation, RNA-seq expression and 5′ peaks information.
The colored lines represent the expression coverage of this region in each condition. Green arrows represent the existing annotation. 5′ peaks are indicated with a vertical red line, blue arrows that follow 5′ peaks denote the potential Transcriptional unit and follows the expression of this region. a. Example of a CDS of Synechococcus (Synpcc7942_0114) with one clear 5′ enriched indication, RNA-seq coverage and proper annotation. b. Example of a TU inside non-coding region with one clear 5′ peak. c. Example of an annotated CDS (Synpcc7942_0174) without 5′ peak and ambiguous RNA-seq coverage. d. Example of a CDS (Synpcc7942_0801) with evidence of structural misannotation since the 5′ peak is located 60 bps after the annotated start site. e. Two annotated genes with contiguous expression and a peak at the 5′ of the first gene suggesting that both CDSs are expressed together as one transcript forming an operon.
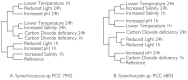
Figure 2. Hierarchical clustering of stress conditions for Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803.
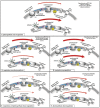
Figure 3. Model: How the relative expression of components of the electron transport chains of photosynthesis and respiration may affect the energy flow at specified time points of stress acclimation.
The thickness of the respective arrows represents the suggested intensity of the energy flow. The conditions that apply to the panel are shown in the upper right of the panel. Panel A. Reference conditions, in the presence of light the photosynthetic electron transport chain has a higher capacity of electron flow than the respiratory chain. Panel B. Conditions where photosynthetic genes are upregulated and respiratory ones are downregulated (Synechoccus: alkaline pH (1-h & 24-h), low light (1-h & 24-h), low temperature (24-h), and Synechocystis: low temperature (24-h)). Panel C. Conditions where respiratory genes are upregulated (_Synechococcu_s: low temperature (24-h) and carbon limitation (1-h)). Panel D. Conditions where photosynthetic and respiratory genes are both downregulated (Synechocystis: low light (1-h & 24-h), elevated salinity (1-h)). Panel E. Conditions where photosynthetic genes are downregulated (Synechocystis: low temperature (1-h), and carbon limitation (24-h)). For simplicity, the two electron transport chains are represented in two different membrane sections, even though respiration is met in the cytoplasmic and both respiration and photosynthesis are intertwined in the thylakoid membrane. The intersection of the two pathways in the thylakoids is more apparent in Synechocystis than Synechococcus, where those two processes are more separated between the cytoplasmic and the thylakoid membrane. This representation only serves the portrayal of the transcriptional changes and their putative affect on energy flow and not the actual distribution of the two chains between the thylakoid and the cytoplasmic membrane. Abbreviations: Temperature (Tm).
Similar articles
- The Flavodiiron Protein Flv3 Functions as a Homo-Oligomer During Stress Acclimation and is Distinct from the Flv1/Flv3 Hetero-Oligomer Specific to the O2 Photoreduction Pathway.
Mustila H, Paananen P, Battchikova N, Santana-Sánchez A, Muth-Pawlak D, Hagemann M, Aro EM, Allahverdiyeva Y. Mustila H, et al. Plant Cell Physiol. 2016 Jul;57(7):1468-1483. doi: 10.1093/pcp/pcw047. Epub 2016 Mar 2. Plant Cell Physiol. 2016. PMID: 26936793 Free PMC article. - Different strategies of metabolic regulation in cyanobacteria: from transcriptional to biochemical control.
Jablonsky J, Papacek S, Hagemann M. Jablonsky J, et al. Sci Rep. 2016 Sep 9;6:33024. doi: 10.1038/srep33024. Sci Rep. 2016. PMID: 27611502 Free PMC article. - Genetic, Genomics, and Responses to Stresses in Cyanobacteria: Biotechnological Implications.
Cassier-Chauvat C, Blanc-Garin V, Chauvat F. Cassier-Chauvat C, et al. Genes (Basel). 2021 Mar 29;12(4):500. doi: 10.3390/genes12040500. Genes (Basel). 2021. PMID: 33805386 Free PMC article. Review. - Cyanobacterial multi-copy chromosomes and their replication.
Watanabe S. Watanabe S. Biosci Biotechnol Biochem. 2020 Jul;84(7):1309-1321. doi: 10.1080/09168451.2020.1736983. Epub 2020 Mar 11. Biosci Biotechnol Biochem. 2020. PMID: 32157949 Review.
Cited by
- Transcriptomic and Phenomic Investigations Reveal Elements in Biofilm Repression and Formation in the Cyanobacterium Synechococcus elongatus PCC 7942.
Simkovsky R, Parnasa R, Wang J, Nagar E, Zecharia E, Suban S, Yegorov Y, Veltman B, Sendersky E, Schwarz R, Golden SS. Simkovsky R, et al. Front Microbiol. 2022 Jun 23;13:899150. doi: 10.3389/fmicb.2022.899150. eCollection 2022. Front Microbiol. 2022. PMID: 35814646 Free PMC article. - Comparative Genomics of the Baltic Sea Toxic Cyanobacteria Nodularia spumigena UHCC 0039 and Its Response to Varying Salinity.
Teikari JE, Hou S, Wahlsten M, Hess WR, Sivonen K. Teikari JE, et al. Front Microbiol. 2018 Mar 8;9:356. doi: 10.3389/fmicb.2018.00356. eCollection 2018. Front Microbiol. 2018. PMID: 29568283 Free PMC article. - Selective molecular transport across the protein shells of bacterial microcompartments.
Bobik TA, Stewart AM. Bobik TA, et al. Curr Opin Microbiol. 2021 Aug;62:76-83. doi: 10.1016/j.mib.2021.05.006. Epub 2021 Jun 1. Curr Opin Microbiol. 2021. PMID: 34087617 Free PMC article. Review. - Comparative Genomic Analysis Revealed Distinct Molecular Components and Organization of CO2-Concentrating Mechanism in Thermophilic Cyanobacteria.
Tang J, Zhou H, Yao D, Riaz S, You D, Klepacz-Smółka A, Daroch M. Tang J, et al. Front Microbiol. 2022 May 6;13:876272. doi: 10.3389/fmicb.2022.876272. eCollection 2022. Front Microbiol. 2022. PMID: 35602029 Free PMC article. - Bacteria can anticipate the seasons: photoperiodism in cyanobacteria.
Jabbur ML, Johnson CH. Jabbur ML, et al. bioRxiv [Preprint]. 2024 Aug 8:2024.05.13.593996. doi: 10.1101/2024.05.13.593996. bioRxiv. 2024. PMID: 38798677 Free PMC article. Updated. Preprint.
References
- Schopf JW (1993) Microfossils of the Early Archean Apex chert: new evidence of the antiquity of life. Science 260: 640–646. - PubMed
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, et al. (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3: 109–136. - PubMed
- Copeland A, Lucas S, Lapidus A, Barry KW, Detter JC, et al.. (n.d.) Complete sequence of chromosome 1 of Synechococcus elongatus PCC 7942. Available: http://www.ncbi.nlm.nih.gov/nuccore/CP000100.1.
Publication types
MeSH terms
Substances
Grants and funding
The U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under contract DE-AC02-05CH11231. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases