Chromosome conformation maps in fission yeast reveal cell cycle dependent sub nuclear structure - PubMed (original) (raw)
. 2014 Nov 10;42(20):12585-99.
doi: 10.1093/nar/gku965. Epub 2014 Oct 23.
Affiliations
- PMID: 25342201
- PMCID: PMC4227791
- DOI: 10.1093/nar/gku965
Chromosome conformation maps in fission yeast reveal cell cycle dependent sub nuclear structure
Ralph S Grand et al. Nucleic Acids Res. 2014.
Abstract
Successful progression through the cell cycle requires spatial and temporal regulation of gene transcript levels and the number, positions and condensation levels of chromosomes. Here we present a high resolution survey of genome interactions in Schizosaccharomyces pombe using synchronized cells to investigate cell cycle dependent changes in genome organization and transcription. Cell cycle dependent interactions were captured between and within S. pombe chromosomes. Known features of genome organization (e.g. the clustering of telomeres and retrotransposon long terminal repeats (LTRs)) were observed throughout the cell cycle. There were clear correlations between transcript levels and chromosomal interactions between genes, consistent with a role for interactions in transcriptional regulation at specific stages of the cell cycle. In silico reconstructions of the chromosome organization within the S. pombe nuclei were made by polymer modeling. These models suggest that groups of genes with high and low, or differentially regulated transcript levels have preferred positions within the S. pombe nucleus. We conclude that the S. pombe nucleus is spatially divided into functional sub-nuclear domains that correlate with gene activity. The observation that chromosomal interactions are maintained even when chromosomes are fully condensed in M phase implicates genome organization in epigenetic inheritance and bookmarking.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Figure 1.
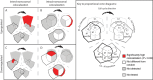
Genome interactions change throughout the Schizosaccharomyces pombe cell cycle. (A) Significant captured interactions between non-adjacent uniquely positioned restriction fragments were tabulated according to whether they were inter- or intrachromosomal (‘Materials and Methods’ section). Most interactions were intrachromosomal, especially following replication and entry into the G2 and M phases. Intersections between the (Bi) inter- and (Bii) intrachromosomal interactions captured in the three cell cycle phases were calculated and plotted as proportional Venn-diagrams in which the area reflects the number of interactions. (Bi) The majority of captured interchromosomal interactions were specific to one cell cycle phase. (Bii) By contrast, the majority of captured intrachromosomal interactions were shared among the three cell cycle phases.
Figure 2.
LTR colocalization predominantly occurs between chromosomes and varies thoughout the cell cycle. (A) The colocalization between LTRs captured in the interactions for each cell cycle phase was determined and the proportion of inter- (light gray) compared to intrachromosomal (dark gray) LTR colocalization was calculated. The majority of LTR colocalization occurs between chromosomes rather than within chromosomes. (Bi) The frequency with which LTR elements from different chromosomes colocalize in the different interchromosomal subsets (Figure 1Bi) was compared to randomly selected genomic regions (see ‘Materials and Methods’ section). LTRs from different chromosomes colocalize at a significantly high frequency in all three phases of the cell cycle. (Bii) Intrachromosomal LTR colocalization also occurred at a significantly high frequency in G1, G2 and M phases, but was not different from random among interactions that were shared by all three cell cycle phases (center) and was not detected in the G2–M phase shared interactions. Individual _P_-values are presented in Supplementary Table S7. The expected false detection rate (FDR) for B(i) and (ii) is 7%.
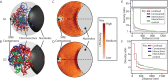
Figure 3.
Polymer models indicate LTRs colocalize close to the nucleolus. (A and B) Cartoons illustrating Schizosaccharomyces pombe genome organization during the: (A) G1; and (B) G2 phases of the cell cycle. Ensembles of coarse-grained polymer models that incorporated biological constraints and captured interactions were generated (Supplementary Methods). Representative models are shown for each cell cycle phase. SPB refers to the spindle pole body. Chromosome I, red; Chromosome II, blue; Chromosome III, green. (C and D) Two-dimensional (2D) projections of LTR density across the ensembles of polymer models show enrichment of LTRs in a nucleolar proximal region during the: (C) G1; and (D) G2 phases of the cell cycle. (E and F) Analyses of the density distribution functions (DDFs) within the: (E) G1; and (F) G2 phases of the cell cycle. Comparisons of the DDFs determined from LTR interactions in ‘constrained’ (tethering restraints only, blue line) or ‘confined’ (spatial confinement only, red line) models indicate significant colocalization (P <0.001; two-tailed, unpaired _t_-test, see Supplementary Methods). This colocalization is not due to a general increase in polymer compaction as colocalization of the LTR elements in the model (‘interactions’, black line) is significantly different (P < 0.001) to that for an equivalent set of random loci (‘interactions control’, green line).
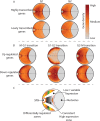
Figure 4.
Genes with high transcript levels form cell cycle-specific interchromosomal clusters. (A) Inter- (light gray) and intrachromosomal (dark gray) colocalization between high transcript genes (Supplementary Table S4, Supplementary Files 4 and 6) within the captured interactions at each phase of the cell cycle was determined. Colocalization between high transcript level genes predominantly occurs between different chromosomes. (B) The frequency with which high transcript genes colocalize in the different interchromosomal subsets (Figure 1Bi) was compared to randomly selected genomic regions (see ‘Materials and Methods’ section). High transcript genes from different chromosomes colocalize at a significantly high frequency in the interactions shared by the M/G1, G1/G2 and G2/M transitions of the cell cycle. (C) Intrachromosomal high transcript gene colocalization also occurred at a significantly high frequency in the G1 phase. By contrast, highly expressed genes colocalized at a significantly low frequency not colocalized in the G1/G2 and G2/M transitions during the G2 phase in interactions that where captured at all cell cycle phases (central blue segment). The high transcript gene set is highly conserved between cell cycle phases (Supplementary Figure S9). (D) The proportion of intrachromosomal colocalization detected between low transcript level genes (Supplementary Table S4, Supplementary Files 4 and 6) was significantly higher than for high transcript genes (_P_-value = <0.05, R: prop.test) resulting in predominant intrachromosomal colocalization. Genes that had no detectable transcripts were excluded from this analysis. Colors as in (A). (**E**) Significant interchromosomal colocalization of low expressed genes was only observed in the G1 phase via interactions that were shared by G1 and G2. (**F**) Despite being responsible for >60% of the observed low transcript colocalization, intrachromosomal colocalization between low transcript genes occurred at or below levels expected at random. Individual _P_-values are presented in Supplementary Table S11. The expected FDR for (B, C, D and E) is 14%.
Figure 5.
Differentially regulated genes colocalize in a cell cycle-specific manner. Genes that had a ≥2-fold change in transcript level during each cell cycle transition were identified (Supplementary File 5). We then determined whether genes with up- or downregulated transcript levels colocalized at a frequency significantly different from randomly selected genomic regions. A significantly high level of (A) inter- and (B) intrachromosomal colocalization was detected between genes that were upregulated during the G1→G2 transition. (C) Interchromosomal colocalization of downregulated genes occurred at a significantly high level in captured interactions that were shared by M and G1 phases of the cell cycle. (D) Intrachromosomal colocalization of downregulated genes occurred at a significantly high level in the M→G1 and G2→M phase transitions. Venn-diagrams were modified to remove the interaction subsets that were not tested during specific cell cycle phase transitions. Individual _P_-values are presented in Supplementary Table S12. The expected FDR for (A–D) is 10%.
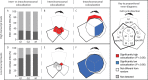
Figure 6.
Highly transcribed and differentially regulated genes have preferred positions in Schizosaccharomyces pombe nuclei. Genes with high, low or differential transcript levels were mapped onto coarse-grained polymer models. The relative densities of the gene sets were averaged over all model ensembles and plotted as 2D gene density maps (Supplementary Methods). Here we present the gene density maps for the interactions models (see Supplementary Figure S11 for gene density maps of the confined and constrained models). The positions of genes that had differential transcript levels during the G1→G2 transition were mapped onto G1 phase polymer models (see Supplementary Figure S12 for the G1→G2 transition gene density mapped on the G2 polymer model). (A) Genes with high transcript levels occupy a central region within the S. pombe nuclei during G1 and G2. By contrast, genes with low transcript levels are dispersed throughout the nucleus. (B) Genes that are upregulated during the M→G1, G1→G2 and G2→M phase transitions occupy distinct nuclear sub-domains. (C) Cartoon highlighting the gross organization of the genes that are predominantly differentially regulated, highly and lowly expressed within the S. pombe nucleus during the G1 and G2 cell cycle phases. SPB: spindle pole body.
Similar articles
- Spatial organization of the Schizosaccharomyces pombe genome within the nucleus.
Matsuda A, Asakawa H, Haraguchi T, Hiraoka Y. Matsuda A, et al. Yeast. 2017 Feb;34(2):55-66. doi: 10.1002/yea.3217. Epub 2016 Nov 29. Yeast. 2017. PMID: 27766670 Review. - Large-scale transcriptome data reveals transcriptional activity of fission yeast LTR retrotransposons.
Mourier T, Willerslev E. Mourier T, et al. BMC Genomics. 2010 Mar 12;11:167. doi: 10.1186/1471-2164-11-167. BMC Genomics. 2010. PMID: 20226011 Free PMC article. - Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation.
Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K. Tanizawa H, et al. Nucleic Acids Res. 2010 Dec;38(22):8164-77. doi: 10.1093/nar/gkq955. Epub 2010 Oct 28. Nucleic Acids Res. 2010. PMID: 21030438 Free PMC article. - The cell cycle-regulated genes of Schizosaccharomyces pombe.
Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, Skiena S, Futcher B, Leatherwood J. Oliva A, et al. PLoS Biol. 2005 Jul;3(7):e225. doi: 10.1371/journal.pbio.0030225. Epub 2005 Jun 28. PLoS Biol. 2005. PMID: 15966770 Free PMC article. - Homologous chromosome pairing in Schizosaccharomyces pombe.
Wells JL, Pryce DW, McFarlane RJ. Wells JL, et al. Yeast. 2006 Oct 15;23(13):977-89. doi: 10.1002/yea.1403. Yeast. 2006. PMID: 17072890 Review.
Cited by
- Data for chromosome contacts and matched transcription profiles at three cell cycle phases in the fission yeast.
Grand RS, O'Sullivan JM. Grand RS, et al. Genom Data. 2015 Jan 20;4:12-6. doi: 10.1016/j.gdata.2015.01.005. eCollection 2015 Jun. Genom Data. 2015. PMID: 26484169 Free PMC article. - Spatial Genome Organization and Its Emerging Role as a Potential Diagnosis Tool.
Meaburn KJ. Meaburn KJ. Front Genet. 2016 Jul 26;7:134. doi: 10.3389/fgene.2016.00134. eCollection 2016. Front Genet. 2016. PMID: 27507988 Free PMC article. Review. - Bioinformatical dissection of fission yeast DNA replication origins.
Masuda K, Renard-Guillet C, Shirahige K, Sutani T. Masuda K, et al. Open Biol. 2020 Jul;10(7):200052. doi: 10.1098/rsob.200052. Epub 2020 Jul 22. Open Biol. 2020. PMID: 32692956 Free PMC article. - Comparative 3D genome structure analysis of the fission and the budding yeast.
Gong K, Tjong H, Zhou XJ, Alber F. Gong K, et al. PLoS One. 2015 Mar 23;10(3):e0119672. doi: 10.1371/journal.pone.0119672. eCollection 2015. PLoS One. 2015. PMID: 25799503 Free PMC article. - Impact of Chromosomal Context on Origin Selection and the Replication Program.
Lanteri L, Perrot A, Schausi-Tiffoche D, Wu PJ. Lanteri L, et al. Genes (Basel). 2022 Jul 14;13(7):1244. doi: 10.3390/genes13071244. Genes (Basel). 2022. PMID: 35886027 Free PMC article.
References
- De Wit E., Bouwman B.a.M., Zhu Y., Klous P., Splinter E., Verstegen M.J.a.M., Krijger P.H.L., Festuccia N., Nora E.P., Welling M., et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases