Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice - PubMed (original) (raw)
Clinical Trial
. 2014 Nov 11;111(45):16106-11.
doi: 10.1073/pnas.1418895111. Epub 2014 Oct 27.
Lorraine Jones-Brando 2, David D Dunigan 3, Geetha Kannan 4, Faith Dickerson 5, Emily Severance 2, Sarven Sabunciyan 2, C Conover Talbot Jr 6, Emese Prandovszky 2, James R Gurnon 3, Irina V Agarkova 3, Flora Leister 2, Kristin L Gressitt 2, Ou Chen 2, Bryan Deuber 2, Fangrui Ma 3, Mikhail V Pletnikov 4, James L Van Etten 7
Affiliations
- PMID: 25349393
- PMCID: PMC4234575
- DOI: 10.1073/pnas.1418895111
Clinical Trial
Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice
Robert H Yolken et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Chloroviruses (family Phycodnaviridae) are large DNA viruses known to infect certain eukaryotic green algae and have not been previously shown to infect humans or to be part of the human virome. We unexpectedly found sequences homologous to the chlorovirus Acanthocystis turfacea chlorella virus 1 (ATCV-1) in a metagenomic analysis of DNA extracted from human oropharyngeal samples. These samples were obtained by throat swabs of adults without a psychiatric disorder or serious physical illness who were participating in a study that included measures of cognitive functioning. The presence of ATCV-1 DNA was confirmed by quantitative PCR with ATCV-1 DNA being documented in oropharyngeal samples obtained from 40 (43.5%) of 92 individuals. The presence of ATCV-1 DNA was not associated with demographic variables but was associated with a modest but statistically significant decrease in the performance on cognitive assessments of visual processing and visual motor speed. We further explored the effects of ATCV-1 in a mouse model. The inoculation of ATCV-1 into the intestinal tract of 9-11-wk-old mice resulted in a subsequent decrease in performance in several cognitive domains, including ones involving recognition memory and sensory-motor gating. ATCV-1 exposure in mice also resulted in the altered expression of genes within the hippocampus. These genes comprised pathways related to synaptic plasticity, learning, memory formation, and the immune response to viral exposure.
Keywords: chlorovirus ATCV-1; cognitive functioning; infection; metagenomic sequencing; oropharyngeal virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
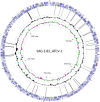
Fig. 1.
Chlorovirus ATCV-1 genome showing the gene block distributions [blue arrows, protein coding sequence (CDS); red arrows, tRNAs] on each strand of the genome. Histograms in black indicate the G+C distribution along the genome; colored histograms (green, magenta) indicate the GC skew of the genome. The most inner circle indicates the genome map position with the start position at “12 o’clock.” The viral genome is a linear dsDNA, but is represented here as a circle for convenience of presentation. Control throat swab deep sequencing consensus sequence reads are matched to ATCV-1, and two experiments (17 and 16 individuals per experiment) are represented by the black lines connecting the gene blocks. BLAST hits, 61; Query, ATCV-1; Subject, human throat swab chlorovirus consensus sequence reads (52).
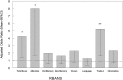
Fig. 2.
Odds of detecting ATCV-1 in the pharynx by percentile of score on cognitive testing. Bars represent the mean and 95% confidence interval odds of detecting ATCV-1 DNA in the oropharynx in individuals with the indicated test. The odds ratios are adjusted for the demographic variables of age, sex, race, maternal education, educational status, and place of birth in the United States. Trails A and Information are separate tests and not part of the RBANS. **P < 0.005, *P < 0.01, adjusted for the same covariates.
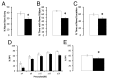
Fig. 3.
Behavioral effects of oral ATCV-1 exposure. Mice were orally infected with C. heliozoae alone (open bars) or with ATCV-1–infected C. heliozoae (solid bars) as described in the text. (A) Spatial recognition memory; the y axis displays the percentage of the previously blocked (i.e., novel) arm entries; *P = 0.015 measured by one-way ANOVA. (B) Novel object recognition; the y axis depicts the percentage of time spent exploring the novel object; *P < 0.001 measured by one-way ANOVA. (C) Place recognition memory recognition; the y axis depicts the percentage of time spent exploring the new location of the familiar object; *P < 0.008 measured by one-way ANOVA. (D) Impaired PPI; mice were exposed to presentation of pulse alone (120 dB) and prepulse–pulse combinations across different prepulse intensities; for example, p4 indicates pairing of the prepulse (4 dB above the background noise of 70 dB) with the pulse alone (120 dB) (see the text for more details); the y axis displays the percentage of PPI. (E) Impaired average PPI; the y axis displays the percentage of PPI; *P < 0.015 measured by post hoc test.
Comment in
- Reply to Kjartansdóttir et al.: Chlorovirus ATCV-1 findings not explained by contamination.
Yolken RH, Jones-Brando L, Dunigan DD, Kannan G, Dickerson F, Severance E, Sabunciyan S, Talbot CC Jr, Prandovszky E, Gurnon JR, Agarkova IV, Leister F, Gressitt KL, Chen O, Deuber B, Ma F, Pletnikov MV, Van Etten JL. Yolken RH, et al. Proc Natl Acad Sci U S A. 2015 Mar 3;112(9):E927. doi: 10.1073/pnas.1424665112. Epub 2015 Feb 5. Proc Natl Acad Sci U S A. 2015. PMID: 25654982 Free PMC article. No abstract available. - Traces of ATCV-1 associated with laboratory component contamination.
Kjartansdóttir KR, Friis-Nielsen J, Asplund M, Mollerup S, Mourier T, Jensen RH, Hansen TA, Rey-Iglesia A, Richter SR, Alquezar-Planas DE, Olsen PV, Vinner L, Fridholm H, Sicheritz-Pontén T, Nielsen LP, Brunak S, Willerslev E, Izarzugaza JM, Hansen AJ. Kjartansdóttir KR, et al. Proc Natl Acad Sci U S A. 2015 Mar 3;112(9):E925-6. doi: 10.1073/pnas.1423756112. Epub 2015 Feb 5. Proc Natl Acad Sci U S A. 2015. PMID: 25654983 Free PMC article. No abstract available.
Similar articles
- Response of Mammalian Macrophages to Challenge with the Chlorovirus Acanthocystis turfacea Chlorella Virus 1.
Petro TM, Agarkova IV, Zhou Y, Yolken RH, Van Etten JL, Dunigan DD. Petro TM, et al. J Virol. 2015 Dec;89(23):12096-107. doi: 10.1128/JVI.01254-15. Epub 2015 Sep 23. J Virol. 2015. PMID: 26401040 Free PMC article. - Reply to Kjartansdóttir et al.: Chlorovirus ATCV-1 findings not explained by contamination.
Yolken RH, Jones-Brando L, Dunigan DD, Kannan G, Dickerson F, Severance E, Sabunciyan S, Talbot CC Jr, Prandovszky E, Gurnon JR, Agarkova IV, Leister F, Gressitt KL, Chen O, Deuber B, Ma F, Pletnikov MV, Van Etten JL. Yolken RH, et al. Proc Natl Acad Sci U S A. 2015 Mar 3;112(9):E927. doi: 10.1073/pnas.1424665112. Epub 2015 Feb 5. Proc Natl Acad Sci U S A. 2015. PMID: 25654982 Free PMC article. No abstract available. - Traces of ATCV-1 associated with laboratory component contamination.
Kjartansdóttir KR, Friis-Nielsen J, Asplund M, Mollerup S, Mourier T, Jensen RH, Hansen TA, Rey-Iglesia A, Richter SR, Alquezar-Planas DE, Olsen PV, Vinner L, Fridholm H, Sicheritz-Pontén T, Nielsen LP, Brunak S, Willerslev E, Izarzugaza JM, Hansen AJ. Kjartansdóttir KR, et al. Proc Natl Acad Sci U S A. 2015 Mar 3;112(9):E925-6. doi: 10.1073/pnas.1423756112. Epub 2015 Feb 5. Proc Natl Acad Sci U S A. 2015. PMID: 25654983 Free PMC article. No abstract available. - Chloroviruses Have a Sweet Tooth.
Van Etten JL, Agarkova I, Dunigan DD, Tonetti M, De Castro C, Duncan GA. Van Etten JL, et al. Viruses. 2017 Apr 22;9(4):88. doi: 10.3390/v9040088. Viruses. 2017. PMID: 28441734 Free PMC article. Review. - Lessons from Chloroviruses: the Complex and Diverse Roles of Viruses in Food Webs.
DeLong JP, Van Etten JL, Dunigan DD. DeLong JP, et al. J Virol. 2023 May 31;97(5):e0027523. doi: 10.1128/jvi.00275-23. Epub 2023 May 1. J Virol. 2023. PMID: 37133447 Free PMC article. Review.
Cited by
- Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria.
Wang W, Jovel J, Halloran B, Wine E, Patterson J, Ford G, OʼKeefe S, Meng B, Song D, Zhang Y, Tian Z, Wasilenko ST, Rahbari M, Reza S, Mitchell T, Jordan T, Carpenter E, Madsen K, Fedorak R, Dielemann LA, Ka-Shu Wong G, Mason AL. Wang W, et al. Inflamm Bowel Dis. 2015 Jun;21(6):1419-27. doi: 10.1097/MIB.0000000000000344. Inflamm Bowel Dis. 2015. PMID: 25939040 Free PMC article. - Functional lability of RNA-dependent RNA polymerases in animals.
Pinzón N, Bertrand S, Subirana L, Busseau I, Escrivá H, Seitz H. Pinzón N, et al. PLoS Genet. 2019 Feb 19;15(2):e1007915. doi: 10.1371/journal.pgen.1007915. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30779744 Free PMC article. - Expression of HLA and Autoimmune Pathway Genes in Liver Biopsies of Young Subjects With Autoimmune Hepatitis Type 1.
Shin E, Schwarz KB, Jones-Brando LV, Florea LD, Sabunciyan S, Wood LD, Yolken RH. Shin E, et al. J Pediatr Gastroenterol Nutr. 2022 Sep 1;75(3):269-275. doi: 10.1097/MPG.0000000000003538. Epub 2022 Jun 27. J Pediatr Gastroenterol Nutr. 2022. PMID: 35759748 Free PMC article. - Contamination Issue in Viral Metagenomics: Problems, Solutions, and Clinical Perspectives.
Jurasz H, Pawłowski T, Perlejewski K. Jurasz H, et al. Front Microbiol. 2021 Oct 20;12:745076. doi: 10.3389/fmicb.2021.745076. eCollection 2021. Front Microbiol. 2021. PMID: 34745046 Free PMC article. Review.
References
- Davari S, Talaei SA, Alaei H, Salami M. Probiotics treatment improves diabetes-induced impairment of synaptic activity and cognitive function: Behavioral and electrophysiological proofs for microbiome-gut-brain axis. Neuroscience. 2013;240:287–296. - PubMed
Publication types
MeSH terms
Grants and funding
- P50 MH094268/MH/NIMH NIH HHS/United States
- MH-94268/MH/NIMH NIH HHS/United States
- P20-RR15635/RR/NCRR NIH HHS/United States
- P20 RR015635/RR/NCRR NIH HHS/United States
- T32 GM008752/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases