Timing and heterogeneity of mutations associated with drug resistance in metastatic cancers - PubMed (original) (raw)
Timing and heterogeneity of mutations associated with drug resistance in metastatic cancers
Ivana Bozic et al. Proc Natl Acad Sci U S A. 2014.
Abstract
Targeted therapies provide an exciting new approach to combat human cancer. The immediate effect is a dramatic reduction in disease burden, but in most cases, the tumor returns as a consequence of resistance. Various mechanisms for the evolution of resistance have been implicated, including mutation of target genes and activation of other drivers. There is increasing evidence that the reason for failure of many targeted treatments is a small preexisting subpopulation of resistant cells; however, little is known about the genetic composition of this resistant subpopulation. Using the novel approach of ordering the resistant subclones according to their time of appearance, here we describe the full spectrum of resistance mutations present in a metastatic lesion. We calculate the expected and median number of cells in each resistant subclone. Surprisingly, the ratio of the medians of successive resistant clones is independent of any parameter in our model; for example, the median of the second clone divided by the median of the first is √2-1. We find that most radiographically detectable lesions harbor at least 10 resistant subclones. Our predictions are in agreement with clinical data on the relative sizes of resistant subclones obtained from liquid biopsies of colorectal cancer patients treated with epidermal growth factor receptor (EGFR) blockade. Our theory quantifies the genetic heterogeneity of resistance that exists before treatment and provides information to design treatment strategies that aim to control resistance.
Keywords: cancer; drug resistance; heterogeneity; mathematical biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
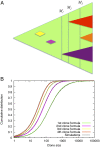
Fig. 1.
Evolution of resistance in a metastatic lesion. (A) As the lesion (green) grows from one cell to detectable size, new resistant subclones appear. Some of them are lost to stochastic drift (yellow and pink), while others survive (purple, red and orange triangle). Instead of looking at the time of appearance of new clones, our approach takes into account the total size of the lesion when the resistance mutation first occurred. (B) Agreement between computer simulations and formula (1) for the cumulative distribution function for the number of cells in the first four resistant clones. The first subclone contains 10 or fewer cells with probability 0.06, between 10 and 100 cells with probability 0.34, between 100 and 1000 cells with probability 0.47 and more than 1000 cells with probability 0.13. The second subclone contains more than 100 cells with probability 0.36. Parameters b=0.25, d=0.181, M=109, u=42⋅10−9.
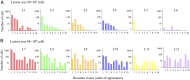
Fig. 2.
Resistant subclones in metastatic lesions. Different realizations of the same stochastic process are shown in each panel. (A) Six lesions of size 108 and (B) six lesions of size 109 cells. The first ten resistant clones are shown, which survived until time of detection. They are ordered according to their time of appearance. Parameter values for all simulations: b=0.25, d=0.181, u=42⋅10−9.
Similar articles
- The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers.
Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B. Diaz LA Jr, et al. Nature. 2012 Jun 28;486(7404):537-40. doi: 10.1038/nature11219. Nature. 2012. PMID: 22722843 Free PMC article. - Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer.
Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, Bencardino K, Cercek A, Chen CT, Veronese S, Zanon C, Sartore-Bianchi A, Gambacorta M, Gallicchio M, Vakiani E, Boscaro V, Medico E, Weiser M, Siena S, Di Nicolantonio F, Solit D, Bardelli A. Misale S, et al. Nature. 2012 Jun 28;486(7404):532-6. doi: 10.1038/nature11156. Nature. 2012. PMID: 22722830 Free PMC article. - The quest to overcome resistance to EGFR-targeted therapies in cancer.
Chong CR, Jänne PA. Chong CR, et al. Nat Med. 2013 Nov;19(11):1389-400. doi: 10.1038/nm.3388. Epub 2013 Nov 7. Nat Med. 2013. PMID: 24202392 Free PMC article. Review. - Strategies to overcome resistance to epidermal growth factor receptor monoclonal antibody therapy in metastatic colorectal cancer.
Jeong WJ, Cha PH, Choi KY. Jeong WJ, et al. World J Gastroenterol. 2014 Aug 7;20(29):9862-71. doi: 10.3748/wjg.v20.i29.9862. World J Gastroenterol. 2014. PMID: 25110417 Free PMC article. Review. - Resistance to EGFR blockade in colorectal cancer: liquid biopsies and latent subclones.
Konieczkowski DJ, Garraway LA. Konieczkowski DJ, et al. Cell Res. 2013 Jan;23(1):13-4. doi: 10.1038/cr.2012.115. Epub 2012 Jul 31. Cell Res. 2013. PMID: 22847744 Free PMC article.
Cited by
- Impact of Resistance on Therapeutic Design: A Moran Model of Cancer Growth.
Lacy MS, Jenner AL. Lacy MS, et al. Bull Math Biol. 2024 Mar 19;86(4):43. doi: 10.1007/s11538-024-01272-6. Bull Math Biol. 2024. PMID: 38502371 Free PMC article. - Citrate cross-feeding by Pseudomonas aeruginosa supports lasR mutant fitness.
Mould DL, Finger CE, Conaway A, Botelho N, Stuut SE, Hogan DA. Mould DL, et al. mBio. 2024 Feb 14;15(2):e0127823. doi: 10.1128/mbio.01278-23. Epub 2024 Jan 23. mBio. 2024. PMID: 38259061 Free PMC article. - Multiscale modeling of drug resistance in glioblastoma with gene mutations and angiogenesis.
Yang H, Lin H, Sun X. Yang H, et al. Comput Struct Biotechnol J. 2023 Oct 21;21:5285-5295. doi: 10.1016/j.csbj.2023.10.037. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37941656 Free PMC article. - Characteristics and molecular mechanism of drug-tolerant cells in cancer: a review.
Liang XW, Liu B, Chen JC, Cao Z, Chu FR, Lin X, Wang SZ, Wu JC. Liang XW, et al. Front Oncol. 2023 Jul 7;13:1177466. doi: 10.3389/fonc.2023.1177466. eCollection 2023. Front Oncol. 2023. PMID: 37483492 Free PMC article. Review. - Mutators can drive the evolution of multi-resistance to antibiotics.
Gifford DR, Berríos-Caro E, Joerres C, Suñé M, Forsyth JH, Bhattacharyya A, Galla T, Knight CG. Gifford DR, et al. PLoS Genet. 2023 Jun 13;19(6):e1010791. doi: 10.1371/journal.pgen.1010791. eCollection 2023 Jun. PLoS Genet. 2023. PMID: 37311005 Free PMC article.
References
- Sawyers C. Targeted cancer therapy. Nature. 2004;432(7015):294–297. - PubMed
- Michor F, et al. Dynamics of chronic myeloid leukaemia. Nature. 2005;435(7046):1267–1270. - PubMed
- Komarova NL, Wodarz D. Targeted Cancer Treatment In Silico: Small Molecule Inhibitors and Oncolytic Viruses. Springer; New York: 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous