COSMIC: exploring the world's knowledge of somatic mutations in human cancer - PubMed (original) (raw)
. 2015 Jan;43(Database issue):D805-11.
doi: 10.1093/nar/gku1075. Epub 2014 Oct 29.
David Beare 2, Prasad Gunasekaran 2, Kenric Leung 2, Nidhi Bindal 2, Harry Boutselakis 2, Minjie Ding 2, Sally Bamford 2, Charlotte Cole 2, Sari Ward 2, Chai Yin Kok 2, Mingming Jia 2, Tisham De 2, Jon W Teague 2, Michael R Stratton 2, Ultan McDermott 2, Peter J Campbell 2
Affiliations
- PMID: 25355519
- PMCID: PMC4383913
- DOI: 10.1093/nar/gku1075
COSMIC: exploring the world's knowledge of somatic mutations in human cancer
Simon A Forbes et al. Nucleic Acids Res. 2015 Jan.
Abstract
COSMIC, the Catalogue Of Somatic Mutations In Cancer (http://cancer.sanger.ac.uk) is the world's largest and most comprehensive resource for exploring the impact of somatic mutations in human cancer. Our latest release (v70; Aug 2014) describes 2 002 811 coding point mutations in over one million tumor samples and across most human genes. To emphasize depth of knowledge on known cancer genes, mutation information is curated manually from the scientific literature, allowing very precise definitions of disease types and patient details. Combination of almost 20,000 published studies gives substantial resolution of how mutations and phenotypes relate in human cancer, providing insights into the stratification of mutations and biomarkers across cancer patient populations. Conversely, our curation of cancer genomes (over 12,000) emphasizes knowledge breadth, driving discovery of unrecognized cancer-driving hotspots and molecular targets. Our high-resolution curation approach is globally unique, giving substantial insight into molecular biomarkers in human oncology. In addition, COSMIC also details more than six million noncoding mutations, 10,534 gene fusions, 61,299 genome rearrangements, 695,504 abnormal copy number segments and 60,119,787 abnormal expression variants. All these types of somatic mutation are annotated to both the human genome and each affected coding gene, then correlated across disease and mutation types.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
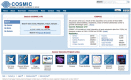
Figure 1.
COSMIC website front page. Search options are presented in the left hand panel, descriptions of the content in the right side panel. The lower panel details related websites and other components of COSMIC. The dark bar at the top provides primary navigation to Help, Downloads and other descriptive content as well as a Contact link to the COSMIC helpdesk. Primary access to COSMIC is via the Search box in the left side panel, accepting multiple parameters including gene names, disease descriptions, mutation syntax and stable COSMIC IDs. ‘Search via Cancer Browser’ raises a new page providing navigation of mutation spectra behind thousands of cancer disease classifications.
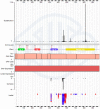
Figure 2.
Full mutation distribution across all tissues and cancer diseases for the KIT gene. The X-axis describes the full length of the gene's coding sequence, and is zoomable (click & drag) to resolve amino acid or nucleotide sequences. In each section of data, the vertical height is kept static, while the scale changes according to the amount of data displayed. From the top down, the following mutation types are displayed: Single base substitutions, gene sequence, PFAM representation of peptide structure, copy number gain (pink)/loss (blue), gene over-(red)/under-(green) expression, multinucleotide substitutions (‘complex’), simple insertions (red triangles) and deletions (blue triangles).
Figure 3.
Example use of filters on the Gene Analysis page to explore disease-specific mutation burden, comparing the mutation profile of the KIT gene in two different tumor types, (A) Hematological and lymphoid: mast cell neoplasm and (B) Soft tissue: gastrointestinal stromal tumor. Substantial differences are very clearly shown in the mutation peaks between the two diseases in this gene, suggesting molecular biomarkers which may be exploited diagnostically, or in pharmaceutical target validation.
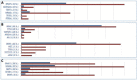
Figure 4.
Histograms from the Cancer Browser, describing the most mutated top few genes on (A) skin melanoma (n = 9136), (B) rare blood cancers hairy cell leukemia (n = 514); Langerhans cell histiocytosis (n = 188) and (C) uveal melanoma (n = 714). Red bars represent the number of samples tested for each gene in the selected disease (‘_n_’), while blue bars represent the number of samples mutated; mutation rates simply calculate n_mutated/n_tested in each case. In this example, BRAF is a well known driver of skin melanoma, mutated in 44% of tumors tested (A). However, BRAF mutations are found at a much higher rate in very restricted populations with rare blood cancers (B). The low mutation frequency of BRAF in uveal (Eye) melanoma (6%) suggests very different genetic mechanisms behind this disease.
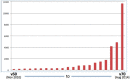
Figure 5.
Growth of COSMIC database size (in Mb, of the Oracle ‘exp’ export file) between November 2010 and August 2014, emphasizing the rapid expansion as COSMIC reflects the data generated by cancer genome studies.
Similar articles
- COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer.
Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, Teague JW, Stratton MR, Futreal PA. Forbes SA, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D652-7. doi: 10.1093/nar/gkp995. Epub 2009 Nov 11. Nucleic Acids Res. 2010. PMID: 19906727 Free PMC article. - COSMIC: the Catalogue Of Somatic Mutations In Cancer.
Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, Harsha B, Hathaway C, Jupe SC, Kok CY, Noble K, Ponting L, Ramshaw CC, Rye CE, Speedy HE, Stefancsik R, Thompson SL, Wang S, Ward S, Campbell PJ, Forbes SA. Tate JG, et al. Nucleic Acids Res. 2019 Jan 8;47(D1):D941-D947. doi: 10.1093/nar/gky1015. Nucleic Acids Res. 2019. PMID: 30371878 Free PMC article. - COSMIC: somatic cancer genetics at high-resolution.
Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ. Forbes SA, et al. Nucleic Acids Res. 2017 Jan 4;45(D1):D777-D783. doi: 10.1093/nar/gkw1121. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899578 Free PMC article. - The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers.
Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. Sondka Z, et al. Nat Rev Cancer. 2018 Nov;18(11):696-705. doi: 10.1038/s41568-018-0060-1. Nat Rev Cancer. 2018. PMID: 30293088 Free PMC article. Review. - Exploring the genomes of cancer cells: progress and promise.
Stratton MR. Stratton MR. Science. 2011 Mar 25;331(6024):1553-8. doi: 10.1126/science.1204040. Science. 2011. PMID: 21436442 Review.
Cited by
- The discrepancy of somatic BRCA1/2 pathogenic variants from two different platforms in epithelial ovarian, fallopian tube, and peritoneal cancer.
Kim JH, Shin JY, Park SY, Seo SS, Kang S, Yoo CW, Park SY, Lim MC. Kim JH, et al. Sci Rep. 2024 Oct 29;14(1):25879. doi: 10.1038/s41598-024-75230-6. Sci Rep. 2024. PMID: 39468117 Free PMC article. - STAT3: Key targets of growth-promoting receptor positive breast cancer.
Jiang RY, Zhu JY, Zhang HP, Yu Y, Dong ZX, Zhou HH, Wang X. Jiang RY, et al. Cancer Cell Int. 2024 Oct 28;24(1):356. doi: 10.1186/s12935-024-03541-9. Cancer Cell Int. 2024. PMID: 39468521 Free PMC article. Review. - Whole-genome sequencing of phenotypically distinct inflammatory breast cancers reveals similar genomic alterations to non-inflammatory breast cancers.
Li X, Kumar S, Harmanci A, Li S, Kitchen RR, Zhang Y, Wali VB, Reddy SM, Woodward WA, Reuben JM, Rozowsky J, Hatzis C, Ueno NT, Krishnamurthy S, Pusztai L, Gerstein M. Li X, et al. Genome Med. 2021 Apr 26;13(1):70. doi: 10.1186/s13073-021-00879-x. Genome Med. 2021. PMID: 33902690 Free PMC article. - Aneuploidy and chromosomal instability in cancer: a jackpot to chaos.
Giam M, Rancati G. Giam M, et al. Cell Div. 2015 May 20;10:3. doi: 10.1186/s13008-015-0009-7. eCollection 2015. Cell Div. 2015. PMID: 26015801 Free PMC article. - Comparative analysis of the mutational landscape and evolutionary patterns of pancreatic ductal adenocarcinoma metastases in the liver or peritoneum.
Yao G, Zhu Y, Liu C, Man Y, Liu K, Zhang Q, Tan Y, Duan Q, Chen D, Du Z, Fan Y. Yao G, et al. Heliyon. 2024 Jul 30;10(15):e35428. doi: 10.1016/j.heliyon.2024.e35428. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39170579 Free PMC article.
References
- Collins F.S., Barker A.D. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci. Am. 2007;296:50–57. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources