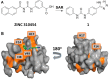Structural analysis of a novel small molecule ligand bound to the CXCL12 chemokine - PubMed (original) (raw)
. 2014 Nov 26;57(22):9693-9.
doi: 10.1021/jm501194p. Epub 2014 Nov 13.
Affiliations
- PMID: 25356720
- PMCID: PMC4255719
- DOI: 10.1021/jm501194p
Structural analysis of a novel small molecule ligand bound to the CXCL12 chemokine
Emmanuel W Smith et al. J Med Chem. 2014.
Abstract
CXCL12 binds to CXCR4, promoting both chemotaxis of lymphocytes and metastasis of cancer cells. We previously identified small molecule ligands that bind CXCL12 and block CXCR4-mediated chemotaxis. We now report a 1.9 Å resolution X-ray structure of CXCL12 bound by such a molecule at a site normally bound by sY21 of CXCR4. The complex structure reveals binding hot spots for future inhibitor design and suggests a new approach to targeting CXCL12-CXCR4 signaling in drug discovery.
Figures
Figure 1
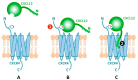
Monomeric representation of CXCR4 bound by CXCL12 through a two-step/two-site process. (A) CXCR4 has a flexible extracellular N-terminal domain. (B) In step-1/site-1, CXCL12 recognizes and binds the N-terminal domain of CXCR4 aided by sulfotyrosine recognition. (C) In step-2/site-2, the flexible N-terminal domain of CXCL12 docks into CXCR4 causing activation. Multiple lines of evidence suggest that CXCR4 can form dimers, but there is no evidence to suggest that the site 1 interface would be altered by a change in oligomeric state of the receptor.
Figure 2
A ZINC 310454 derivative binds in the sY21 binding pocket as predicted by in silico docking. (A) Fragment-based SAR analysis of ZINC 310454 led to the design and synthesis of compound 1. (B) Significant chemical shift perturbations in the presence of 1600 μM compound 1 map to the predicted binding pocket on CXCL12 (PDB: 2K05). The residues most perturbed are colored in orange.
Figure 3
Complex crystallographic structure of CXCL12 dimer (ribbon and surface model) with compound 1 (yellow) bound to the sY21-binding site.
Figure 4
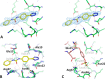
Complex crystallographic structure and characterization of the sY21 binding site. (A) Stereo image depiction of the unbiased _F_o−_F_c map (at 3σ) of compound 1 bound to the sY21-binding site of CXCL12. (B) Complex crystal structure of CXCL12 (green) bound by compound 1 (yellow) superimposed to the apo crystal structure (PDB ID: 2J7Z) (cyan) shows conformational changes induced upon binding. (C) NMR complex structure of CXCL12 bound to the D20-sY21-D22 segment of CXCR41–38 (PDB ID: 2K05) outlines the sY21-binding site as well as the possible hydrogen bond interactions.
Figure 5
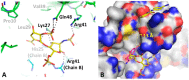
Heparin/sY12 vs compound 1 binding. (A) CXCL12–heparin sulfate crystal structure (PDB ID: 2NWG) positions haparin sulfate (yellow) in the region where sY12 of CXCR41–38 normally binds. (B) CXCL12–compound 1 crystal structure with heparin sulfate (yellow) and sY12 (purple) superimposed in sY12 site suggests that compounds specific to both sites could potentially be linked together via linkers.
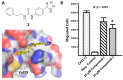
Figure 6
Docking pose and chemotaxis inhibition. (A) Compound 2 is based on the compound 1 scaffold but contains an additional two-carbon linker past the urea. Docking pose prediction suggests the flexible linker may help increase hydrophobic interactions with the cleft above Val39. (B) Chemotaxis assay in THP-1 cells demontrates that 25 μM compound 1 reduces cell migration by ∼20%, while 25 μM compound 2 reduces cell migration by ∼40%.
Similar articles
- Fragment-based optimization of small molecule CXCL12 inhibitors for antagonizing the CXCL12/CXCR4 interaction.
Ziarek JJ, Liu Y, Smith E, Zhang G, Peterson FC, Chen J, Yu Y, Chen Y, Volkman BF, Li R. Ziarek JJ, et al. Curr Top Med Chem. 2012;12(24):2727-40. doi: 10.2174/1568026611212240003. Curr Top Med Chem. 2012. PMID: 23368099 Free PMC article. - Targeting SDF-1/CXCL12 with a ligand that prevents activation of CXCR4 through structure-based drug design.
Veldkamp CT, Ziarek JJ, Peterson FC, Chen Y, Volkman BF. Veldkamp CT, et al. J Am Chem Soc. 2010 Jun 2;132(21):7242-3. doi: 10.1021/ja1002263. J Am Chem Soc. 2010. PMID: 20459090 Free PMC article. - Interaction of chemokine receptor CXCR4 in monomeric and dimeric state with its endogenous ligand CXCL12: coarse-grained simulations identify differences.
Cutolo P, Basdevant N, Bernadat G, Bachelerie F, Ha-Duong T. Cutolo P, et al. J Biomol Struct Dyn. 2017 Feb;35(2):399-412. doi: 10.1080/07391102.2016.1145142. Epub 2016 Jul 11. J Biomol Struct Dyn. 2017. PMID: 26813575 - The good and bad faces of the CXCR4 chemokine receptor.
Teixidó J, Martínez-Moreno M, Díaz-Martínez M, Sevilla-Movilla S. Teixidó J, et al. Int J Biochem Cell Biol. 2018 Feb;95:121-131. doi: 10.1016/j.biocel.2017.12.018. Epub 2017 Dec 27. Int J Biochem Cell Biol. 2018. PMID: 29288743 Review. - Biological/pathological functions of the CXCL12/CXCR4/CXCR7 axes in the pathogenesis of bladder cancer.
Nazari A, Khorramdelazad H, Hassanshahi G. Nazari A, et al. Int J Clin Oncol. 2017 Dec;22(6):991-1000. doi: 10.1007/s10147-017-1187-x. Epub 2017 Oct 11. Int J Clin Oncol. 2017. PMID: 29022185 Review.
Cited by
- Trisubstituted 1,3,5-Triazines: The First Ligands of the sY12-Binding Pocket on Chemokine CXCL12.
Sprague DJ, Getschman AE, Fenske TG, Volkman BF, Smith BC. Sprague DJ, et al. ACS Med Chem Lett. 2021 Oct 18;12(11):1773-1782. doi: 10.1021/acsmedchemlett.1c00388. eCollection 2021 Nov 11. ACS Med Chem Lett. 2021. PMID: 34795867 Free PMC article. - What Do Structures Tell Us About Chemokine Receptor Function and Antagonism?
Kufareva I, Gustavsson M, Zheng Y, Stephens BS, Handel TM. Kufareva I, et al. Annu Rev Biophys. 2017 May 22;46:175-198. doi: 10.1146/annurev-biophys-051013-022942. Annu Rev Biophys. 2017. PMID: 28532213 Free PMC article. Review. - CCR7 Sulfotyrosine Enhances CCL21 Binding.
Phillips AJ, Taleski D, Koplinski CA, Getschman AE, Moussouras NA, Richard AM, Peterson FC, Dwinell MB, Volkman BF, Payne RJ, Veldkamp CT. Phillips AJ, et al. Int J Mol Sci. 2017 Aug 25;18(9):1857. doi: 10.3390/ijms18091857. Int J Mol Sci. 2017. PMID: 28841151 Free PMC article. - Crystallographic Structure of Truncated CCL21 and the Putative Sulfotyrosine-Binding Site.
Smith EW, Lewandowski EM, Moussouras NA, Kroeck KG, Volkman BF, Veldkamp CT, Chen Y. Smith EW, et al. Biochemistry. 2016 Oct 11;55(40):5746-5753. doi: 10.1021/acs.biochem.6b00304. Epub 2016 Sep 26. Biochemistry. 2016. PMID: 27617343 Free PMC article. - Structural Insights into Molecular Recognition by Human Chemokine CCL19.
Lewandowski EM, Kroeck KG, Jacobs LMC, Fenske TG, Witt RN, Hintz AM, Ramsden ER, Zhang X, Peterson F, Volkman BF, Veldkamp CT, Chen Y. Lewandowski EM, et al. Biochemistry. 2022 Mar 1;61(5):311-318. doi: 10.1021/acs.biochem.1c00759. Epub 2022 Feb 14. Biochemistry. 2022. PMID: 35156805 Free PMC article.
References
- Rossi D.; Zlotnik A. The biology of chemokines and their receptors. Annu. Rev. Immunol. 2000, 18, 217–242. - PubMed
- Comerford I.; McColl S. R. Mini-review series: focus on chemokines. Immunol. Cell Biol. 2011, 89, 183–184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM097381/GM/NIGMS NIH HHS/United States
- R37 AI058072/AI/NIAID NIH HHS/United States
- CA173056/CA/NCI NIH HHS/United States
- AI058072/AI/NIAID NIH HHS/United States
- GM097381/GM/NIGMS NIH HHS/United States
- R01 CA173056/CA/NCI NIH HHS/United States
- R01 AI058072/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources