Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly - PubMed (original) (raw)
Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly
Amanda J Price et al. PLoS Pathog. 2014.
Abstract
The HIV-1 capsid is involved in all infectious steps from reverse transcription to integration site selection, and is the target of multiple host cell and pharmacologic ligands. However, structural studies have been limited to capsid monomers (CA), and the mechanistic basis for how these ligands influence infection is not well understood. Here we show that a multi-subunit interface formed exclusively within CA hexamers mediates binding to linear epitopes within cellular cofactors NUP153 and CPSF6, and is competed for by the antiretroviral compounds PF74 and BI-2. Each ligand is anchored via a shared phenylalanine-glycine (FG) motif to a pocket within the N-terminal domain of one monomer, and all but BI-2 also make essential interactions across the N-terminal domain: C-terminal domain (NTD:CTD) interface to a second monomer. Dissociation of hexamer into CA monomers prevents high affinity interaction with CPSF6 and PF74, and abolishes binding to NUP153. The second interface is conformationally dynamic, but binding of NUP153 or CPSF6 peptides is accommodated by only one conformation. NUP153 and CPSF6 have overlapping binding sites, but each makes unique CA interactions that, when mutated selectively, perturb cofactor dependency. These results reveal that multiple ligands share an overlapping interface in HIV-1 capsid that is lost upon viral disassembly.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Binding of HIV-1 hexamer to CPSF6, NUP153, PF74 and BI-2.
(A) ITC binding isotherms of HIV-1 hexamer titrated into different ligands in either the presence or absence of DTT. For both CPSF6 and NUP153, a peptide corresponding to residues 313–327 or 1407–1423 respectively was used. (B) Titration of HIV-1 cofactors CPSF6 and NUP153 into hexamer in the presence of pharmacologic inhibitors. Thermodynamic parameters are indicated for interactions displaying a clear binding isotherm; approximate affinities are shown for weak interactions; NB = no binding detectable.
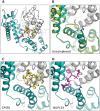
Figure 2. Crystal structures of CPSF6313–327 (A) and NUP1531407–1423 (B) FG-containing peptides in complex with HIV-1 hexamer.
The P6 crystal form is shown, with the six subunits of the HIV-1 capsid generated by crystallographic symmetry. The capsid is shown in a ribbon representation with each monomer colored differently. Atoms comprising the cofactor peptides are shown as spheres, with each peptide colored separately. There are six cofactor binding sites per hexamer and six bound cofactor peptides. For each structure two views are shown, related by 90°.
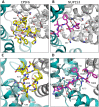
Figure 3. CPSF6 and NUP153 bind a multi-subunit interface in HIV-1 hexamer.
(A) Two monomers from the CPSF6 P212121 complexed structure are shown in gray and teal, with the CPSF6 peptide in yellow and the NUP153 peptide from the P212121 complexed structure superposed and shown in pink. The helices that comprise the binding site are numbered. (B) Two monomers from the uncomplexed P212121 hexamer structure (3H4E) showing representative helices 8 and 9 in ‘open’ (teal) and ‘closed’ (green) states are superposed. The side-chain of residue Q179 is shown in stick representation. (C) CPSF6 binding site in the P212121 structure, with the peptide shown in yellow and subunits colored as above. (D) NUP153 binding site in the P212121 structure, with peptide shown in pink and subunits colored as above. CPSF6 and NUP153 preferentially bind an open conformation of helices 8 and 9.
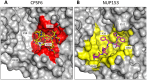
Figure 4. CPSF6 and NUP153 interact with distinct sets of CA residues within the hexamer interface.
A molecular surface representation of the CPSF6 and NUP153 binding site in hexameric capsid is shown in gray. The interface is formed between two neighbouring monomers and is shown in a similar orientation to Figure 3. The CPSF6 (A) and NUP153 (B) peptides in the different complexed structures are shown in yellow and pink respectively. The binding ‘footprint’ of each ligand is shown in red for CPSF6 and yellow for NUP153. The footprint was calculated using PISA and is defined as residues containing atoms that become desolvated upon binding (i.e. are within 1.4 Å of the ligand).
Figure 5. CPSF6 and NUP153 interact both within and between CA monomers.
Detailed views of the interactions between the ligands and the binding site within hexameric capsid are shown. In each case two adjacent monomers of the hexamer are colored gray and teal, with the side chains of specific contacting residues displayed and labelled in standard text. Potential hydrogen bonds with ligands are indicated by dashed lines. The upper panels (A and B) focus on interactions that occur in the first binding site (within one monomer), while the lower panels (C and D), show interactions in the second binding site (with the second monomer). CPSF6 is shown in yellow and NUP153 in pink, with important ligand residues labelled in bold and italic text.
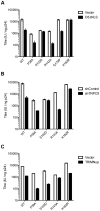
Figure 6. Sensitivity of CA mutants to restriction by CPSF6ΔNLS or TRIM-NUP153 or depletion of TNPO3.
Titres of VSV-G pseudotyped GFP-encoding HIV-1 vectors bearing wild type or mutant CA on HeLa cells expressing empty vector (EV) or CPSF6ΔNLS (A), control knockdown cells (shControl) or cells depleted of TNPO3 (shTNPO3) (B), control knockdown cells (shControl) or cells expressing empty vector (EV) or TRIM-NUP153 (C). In each case infectivity is plotted as infectious units (IU)/ng p24. The data are representative of two independent experiments, each using three different virus doses.
Figure 7. HIV-1 CA hexamer complexes with pharmacologic ligands BI-2 and PF74.
Two monomers from hexameric structures complexed with BI-2 (A) and PF74 (B) are shown in a close-up view of the binding site, with one monomer in gray and a second monomer in teal. BI-2 is shown in orange and PF74 in green. Important CA interacting residues are shown as sticks and putative hydrogen bonds indicated by dashed lines. Ligplots are shown for BI-2 (C) and PF74 (D) in which a ball-and-stick representation of the ligand is shown together with CA residues that contribute a hydrogen bond. Other interacting CA residues are indicated with a red-dashed semi-circle. Circled areas indicate interacting CA residues that are shared between both structures. (E) Superposition of the BI-2:hexamer complex (BI-2 in orange) with the previously solved BI-1:CA-NTD complex (BI-1 in blue). The secondary structure of the superposed monomers from each structure is shown in gray, while capsid side-chains from only the hexamer complex are depicted. The adjacent monomer in the hexamer complex is shown in teal. (F) Superposition of the PF74:hexamer complex (PF74 in green) with the previously solved PF74:CA-NTD complex (PF74 in slate). The capsids are colored as in (E).
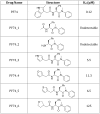
Figure 8. Structures of PF74 derivatives used in this study and their affinities of binding to CA.
Affinities of binding to hexamer as measured by ITC are shown. No binding was detected between PF74_1 and PF74_2 to HIV-1 CA NTD, therefore the interaction with hexamer was not tested.
Figure 9. Inhibition of HIV-1 infection by BI-2, PF74 and PF74 derivatives.
(A) Titration of PF74 or indicated derivatives onto HeLa cells infected with VSV-G pseudotyped GFP-encoding HIV-1 vector. For each titration, infectivity is normalised to the level in the absence of inhibitor (100%). (B) Correlation between the IC90 values derived from (A) and the Kd values for purified hexameric capsid as calculated by ITC (Figure 8). The values are highly correlative with a Pearson correlation coefficient of 0.9928 and a P-value of 0.0007. (C) Titration of PF74 or BI-2 onto cells infected with VSV-G pseudotyped GFP-encoding HIV-1 vectors, normalized as in (A). (D) Data from (C) but with infectivity plotted against drug concentration divided by affinity to hexamer as calculated by ITC. (E) Titres of VSV-G pseudotyped GFP-encoding HIV-1 vectors 48 h post-infection and levels of reverse transcription (RT) 4 h post-infection under conditions of PF74 inhibition. For both measures, data are normalized to the values obtained in the absence of inhibitor. (F) Data from (E), except plotted against the calculated level of CA occupancy by PF74. The occupancy was calculated assuming that there are 1500 free sites per capsid and that number of free sites = 1500(1−([PF74]/Kd[PF74])) (G) Infectivity and reverse transcription in cells infected in the presence of PF74 (wash) or when removed after four hours (wash out). Reverse transcription was then measured after a further 4 h, while infectivity was determined 48 h post-infection. (H) Levels of reverse transcription 4 h post-infection of HeLa cells in the presence of PF74 and its derivatives at 20× IC9o drug concentrations.
Similar articles
- Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.
Bhattacharya A, Alam SL, Fricke T, Zadrozny K, Sedzicki J, Taylor AB, Demeler B, Pornillos O, Ganser-Pornillos BK, Diaz-Griffero F, Ivanov DN, Yeager M. Bhattacharya A, et al. Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):18625-30. doi: 10.1073/pnas.1419945112. Epub 2014 Dec 17. Proc Natl Acad Sci U S A. 2014. PMID: 25518861 Free PMC article. - Roles of Capsid-Interacting Host Factors in Multimodal Inhibition of HIV-1 by PF74.
Saito A, Ferhadian D, Sowd GA, Serrao E, Shi J, Halambage UD, Teng S, Soto J, Siddiqui MA, Engelman AN, Aiken C, Yamashita M. Saito A, et al. J Virol. 2016 May 27;90(12):5808-5823. doi: 10.1128/JVI.03116-15. Print 2016 Jun 15. J Virol. 2016. PMID: 27076642 Free PMC article. - Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells.
Buffone C, Martinez-Lopez A, Fricke T, Opp S, Severgnini M, Cifola I, Petiti L, Frabetti S, Skorupka K, Zadrozny KK, Ganser-Pornillos BK, Pornillos O, Di Nunzio F, Diaz-Griffero F. Buffone C, et al. J Virol. 2018 Sep 12;92(19):e00648-18. doi: 10.1128/JVI.00648-18. Print 2018 Oct 1. J Virol. 2018. PMID: 29997211 Free PMC article. - HIV Capsid and Integration Targeting.
Engelman AN. Engelman AN. Viruses. 2021 Jan 18;13(1):125. doi: 10.3390/v13010125. Viruses. 2021. PMID: 33477441 Free PMC article. Review. - The HIV-1 Capsid: From Structural Component to Key Factor for Host Nuclear Invasion.
Scoca V, Di Nunzio F. Scoca V, et al. Viruses. 2021 Feb 10;13(2):273. doi: 10.3390/v13020273. Viruses. 2021. PMID: 33578999 Free PMC article. Review.
Cited by
- Chromatin organization at the nuclear pore favours HIV replication.
Lelek M, Casartelli N, Pellin D, Rizzi E, Souque P, Severgnini M, Di Serio C, Fricke T, Diaz-Griffero F, Zimmer C, Charneau P, Di Nunzio F. Lelek M, et al. Nat Commun. 2015 Mar 6;6:6483. doi: 10.1038/ncomms7483. Nat Commun. 2015. PMID: 25744187 Free PMC article. - The HIV capsid mimics karyopherin engagement of FG-nucleoporins.
Dickson CF, Hertel S, Tuckwell AJ, Li N, Ruan J, Al-Izzi SC, Ariotti N, Sierecki E, Gambin Y, Morris RG, Towers GJ, Böcking T, Jacques DA. Dickson CF, et al. Nature. 2024 Feb;626(8000):836-842. doi: 10.1038/s41586-023-06969-7. Epub 2024 Jan 24. Nature. 2024. PMID: 38267582 Free PMC article. - HIV-1 Capsid Core: A Bullet to the Heart of the Target Cell.
Toccafondi E, Lener D, Negroni M. Toccafondi E, et al. Front Microbiol. 2021 Apr 1;12:652486. doi: 10.3389/fmicb.2021.652486. eCollection 2021. Front Microbiol. 2021. PMID: 33868211 Free PMC article. Review. - In Vitro Quantification of the Effects of IP6 and Other Small Polyanions on Immature HIV-1 Particle Assembly and Core Stability.
Dostálková A, Kaufman F, Křížová I, Vokatá B, Ruml T, Rumlová M. Dostálková A, et al. J Virol. 2020 Sep 29;94(20):e00991-20. doi: 10.1128/JVI.00991-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32727872 Free PMC article. - HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains.
Francis AC, Marin M, Singh PK, Achuthan V, Prellberg MJ, Palermino-Rowland K, Lan S, Tedbury PR, Sarafianos SG, Engelman AN, Melikyan GB. Francis AC, et al. Nat Commun. 2020 Jul 14;11(1):3505. doi: 10.1038/s41467-020-17256-8. Nat Commun. 2020. PMID: 32665593 Free PMC article.
References
- Franke EK, Yuan HE, Luban J (1994) Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372: 359–362. - PubMed
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, et al. (2008) Identification of host proteins required for HIV infection through a functional genomic screen. Science 319: 921–926. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical