PTMcode v2: a resource for functional associations of post-translational modifications within and between proteins - PubMed (original) (raw)
. 2015 Jan;43(Database issue):D494-502.
doi: 10.1093/nar/gku1081. Epub 2014 Oct 31.
Affiliations
- PMID: 25361965
- PMCID: PMC4383916
- DOI: 10.1093/nar/gku1081
PTMcode v2: a resource for functional associations of post-translational modifications within and between proteins
Pablo Minguez et al. Nucleic Acids Res. 2015 Jan.
Abstract
The post-translational regulation of proteins is mainly driven by two molecular events, their modification by several types of moieties and their interaction with other proteins. These two processes are interdependent and together are responsible for the function of the protein in a particular cell state. Several databases focus on the prediction and compilation of protein-protein interactions (PPIs) and no less on the collection and analysis of protein post-translational modifications (PTMs), however, there are no resources that concentrate on describing the regulatory role of PTMs in PPIs. We developed several methods based on residue co-evolution and proximity to predict the functional associations of pairs of PTMs that we apply to modifications in the same protein and between two interacting proteins. In order to make data available for understudied organisms, PTMcode v2 (http://ptmcode.embl.de) includes a new strategy to propagate PTMs from validated modified sites through orthologous proteins. The second release of PTMcode covers 19 eukaryotic species from which we collected more than 300,000 experimentally verified PTMs (>1,300,000 propagated) of 69 types extracting the post-translational regulation of >100,000 proteins and >100,000 interactions. In total, we report 8 million associations of PTMs regulating single proteins and over 9.4 million interplays tuning PPIs.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
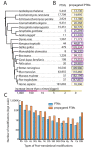
Figure 1.
The PTMcode data set. We collected experimentally verified PTMs from 19 eukaryotes (A) and spread their annotation through conserved sites in orthologs tagging the new annotated sites as ‘propagated PTMs’. (B) Shows the increase on the size of available post-translational information per species. Organisms with fewer validated PTMs are the ones showing a higher increase as shown by the color code. (C) Shows the increment of the most abundant PTM types.
Figure 2.
Propagated PTMs. Users can choose to display the propagated PTMs by clicking in the checkbox in the display menu (A). Here, we show the protein Sox2 in Pan troglodytes (A) which had no PTMs experimentally validated while several are predicted from orthologs, mainly acetylations from the mouse copy of the protein (B) and phosphorylations from human (C). Those PTMs were also propagated between mouse and human (not displayed in the figure).
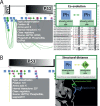
Figure 3.
Co-evolution and structural distance channels. The majority of the functional associations between PTM pairs are found by these two types of evidences. (A) The ‘co-evolution’ channel extracts pairs of PTMs that have a common evolutionary history. The protein MSA used to calculate the co-evolution score is displayed using the Jalview plugin where the two modified residues are highlighted for their closer analysis. (B) The ‘structural distance’ channels extracts PTM pairs that are close in PDB protein structures. The Jmol plugin offers further possibilities for their analysis.
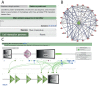
Figure 4.
Visualization of functional associations between PTMs in interacting proteins. Users can explore the regulation of the interactions where their favorite protein is involved (A). We implemented a network viewer where all its interactions are displayed (B). Every partner in the network opens the ‘PPI view’, a framework to explore the functional associations found between PTMs from both proteins (C).
Similar articles
- PTMcode: a database of known and predicted functional associations between post-translational modifications in proteins.
Minguez P, Letunic I, Parca L, Bork P. Minguez P, et al. Nucleic Acids Res. 2013 Jan;41(Database issue):D306-11. doi: 10.1093/nar/gks1230. Epub 2012 Nov 28. Nucleic Acids Res. 2013. PMID: 23193284 Free PMC article. - Bioinformatics Analysis of Functional Associations of PTMs.
Minguez P, Bork P. Minguez P, et al. Methods Mol Biol. 2017;1558:303-320. doi: 10.1007/978-1-4939-6783-4_14. Methods Mol Biol. 2017. PMID: 28150244 - dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins.
Huang KY, Su MG, Kao HJ, Hsieh YC, Jhong JH, Cheng KH, Huang HD, Lee TY. Huang KY, et al. Nucleic Acids Res. 2016 Jan 4;44(D1):D435-46. doi: 10.1093/nar/gkv1240. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578568 Free PMC article. - Mass spectrometric analysis of post-translational modifications (PTMs) and protein-protein interactions (PPIs).
Ngounou Wetie AG, Woods AG, Darie CC. Ngounou Wetie AG, et al. Adv Exp Med Biol. 2014;806:205-35. doi: 10.1007/978-3-319-06068-2_9. Adv Exp Med Biol. 2014. PMID: 24952184 Review. - Software eyes for protein post-translational modifications.
Na S, Paek E. Na S, et al. Mass Spectrom Rev. 2015 Mar-Apr;34(2):133-47. doi: 10.1002/mas.21425. Epub 2014 Jun 2. Mass Spectrom Rev. 2015. PMID: 24889695 Review.
Cited by
- A Review of Machine Learning and Algorithmic Methods for Protein Phosphorylation Site Prediction.
Esmaili F, Pourmirzaei M, Ramazi S, Shojaeilangari S, Yavari E. Esmaili F, et al. Genomics Proteomics Bioinformatics. 2023 Dec;21(6):1266-1285. doi: 10.1016/j.gpb.2023.03.007. Epub 2023 Oct 19. Genomics Proteomics Bioinformatics. 2023. PMID: 37863385 Free PMC article. Review. - Spatiotemporal variation of mammalian protein complex stoichiometries.
Ori A, Iskar M, Buczak K, Kastritis P, Parca L, Andrés-Pons A, Singer S, Bork P, Beck M. Ori A, et al. Genome Biol. 2016 Mar 14;17:47. doi: 10.1186/s13059-016-0912-5. Genome Biol. 2016. PMID: 26975353 Free PMC article. - Analysis and review of techniques and tools based on machine learning and deep learning for prediction of lysine malonylation sites in protein sequences.
Ramazi S, Tabatabaei SAH, Khalili E, Nia AG, Motarjem K. Ramazi S, et al. Database (Oxford). 2024 Jan 19;2024:baad094. doi: 10.1093/database/baad094. Database (Oxford). 2024. PMID: 38245002 Free PMC article. Review. - Kinome-wide identification of phosphorylation networks in eukaryotic proteomes.
Parca L, Ariano B, Cabibbo A, Paoletti M, Tamburrini A, Palmeri A, Ausiello G, Helmer-Citterich M. Parca L, et al. Bioinformatics. 2019 Feb 1;35(3):372-379. doi: 10.1093/bioinformatics/bty545. Bioinformatics. 2019. PMID: 30016513 Free PMC article. - Pathological implication of protein post-translational modifications in cancer.
Pan S, Chen R. Pan S, et al. Mol Aspects Med. 2022 Aug;86:101097. doi: 10.1016/j.mam.2022.101097. Epub 2022 Apr 7. Mol Aspects Med. 2022. PMID: 35400524 Free PMC article. Review.
References
- Yamauchi M., Shiiba M. Lysine hydroxylation and cross-linking of collagen. Methods Mol. Biol. 2008;446:95–108. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous