Nuclear position dictates DNA repair pathway choice - PubMed (original) (raw)
. 2014 Nov 15;28(22):2450-63.
doi: 10.1101/gad.248369.114. Epub 2014 Nov 3.
Anastazja Grabarz 1, Katerina Tsouroula 1, Leonid Andronov 1, Audrey Furst 1, Tibor Pankotai 1, Vincent Heyer 1, Mélanie Rogier 1, Kathleen M Attwood 2, Pascal Kessler 1, Graham Dellaire 2, Bruno Klaholz 1, Bernardo Reina-San-Martin 1, Evi Soutoglou 1
Affiliations
- PMID: 25366693
- PMCID: PMC4233239
- DOI: 10.1101/gad.248369.114
Nuclear position dictates DNA repair pathway choice
Charlène Lemaître et al. Genes Dev. 2014.
Abstract
Faithful DNA repair is essential to avoid chromosomal rearrangements and promote genome integrity. Nuclear organization has emerged as a key parameter in the formation of chromosomal translocations, yet little is known as to whether DNA repair can efficiently occur throughout the nucleus and whether it is affected by the location of the lesion. Here, we induce DNA double-strand breaks (DSBs) at different nuclear compartments and follow their fate. We demonstrate that DSBs induced at the nuclear membrane (but not at nuclear pores or nuclear interior) fail to rapidly activate the DNA damage response (DDR) and repair by homologous recombination (HR). Real-time and superresolution imaging reveal that DNA DSBs within lamina-associated domains do not migrate to more permissive environments for HR, like the nuclear pores or the nuclear interior, but instead are repaired in situ by alternative end-joining. Our results are consistent with a model in which nuclear position dictates the choice of DNA repair pathway, thus revealing a new level of regulation in DSB repair controlled by spatial organization of DNA within the nucleus.
Keywords: DNA repair; alternative end-joining; nuclear lamina; nuclear organization.
© 2014 Lemaître et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
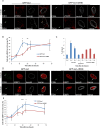
Figure 1.
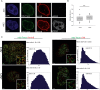
The DDR is delayed at the nuclear lamina. (A) Immuno-FISH single-Z confocal images of the lacO array (green), γ-H2AX (red), and laminB (gray) in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD and treated or not with Dox for 14 h. (B) Time course of the percentage of colocalization of the lacO array with γ-H2AX. (C) γ-H2AX ChIP at the indicated time points after Dox addition in cells expressing GFP-lacI or GFP-lacI-ΔEMD. Values were normalized to input DNA and H3 ChIP and are representative of three independent experiments. (D) Immuno-FISH single-Z confocal images of the lacO array (green), 53BP1 (red), and laminB (gray) in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD and treated or not with Dox for 20 h. (E) 53BP1 after Dox addition in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD. Values represent mean ± SD of three independent experiments with n > 50 cells. For statistical analysis, a _t_-test was performed. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. In all figures, the arrow depicts the position of the lacO array.
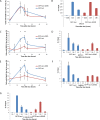
Figure 2.
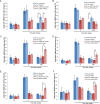
Recruitment of HR factors is impaired at the nuclear lamina. (A) Time course of the percentage of colocalization of the lacO array with Ku80 after Dox addition in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD. Values represent mean ± SD of three independent experiments with n > 50 cells. ChIP for XRRC4 (B), BRCA1 (D), RAD51 (F), or P-RPAS33 (G) at the indicated times upon Dox addition in I-Hela111 cells (XRCC4) or I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD is shown. Values were normalized to input DNA and are representative of three independent experiments. The percentage of colocalization of the lacO array with BRCA1 (C) and Rad51 (E) at the indicated times after Dox addition in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD is shown. Values represent mean ± SD of three independent experiments with n > 50 cells. For statistical analysis, a _t_-test was performed. (*) P < 0.05; (**) P < 0.01.
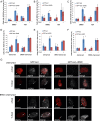
Figure 3.
Chromatin decompaction restores DDR and the recruitment of HR factors at the nuclear lamina. Colocalization of the lacO array with γ-H2AX (A), BRCA1 (B), or RAD51 (C) in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD and pretreated for 4 h with DMSO or TSA in the absence or presence of Dox for 14 h or 20 h is shown. The percentage of colocalization of the lacO array with γ-H2AX (D), BRCA1 (E), or RAD51 (F) in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD and cherry-LacI or BRG1-cherry-lacI and treated or not with Dox for 14 h or 20 h is shown. (G) Immunofluorescence single-Z confocal images of γ-H2AX (gray) in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD transfected with cherry-lacI or BRG1-cherry-lacI (red) and treated or not with Dox for 14 h. For statistical analysis, a _t_-test was performed. (*) P < 0.05; (**) P < 0.01.
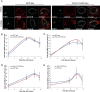
Figure 4.
DDR and HR are not affected by tethering at the nuclear pores. (A) Immuno-FISH single-Z confocal images of the lacO array (green), γ-H2AX (red), and laminB (gray) in I-U2OS19 cells expressing GFP-lacI or Pom121-GFP-lacI and treated or not with Dox for 14 h. Time course of the percentage of colocalization of the lacO array with γ-H2AX (B), 53BP1 (C), BRCA1 (D), or RAD51 (E) in I-U2OS19 cells expressing GFP-lacI or Pom121-GFP-lacI cells after Dox addition is shown. Values represent mean ± SD of three independent experiments with n > 50 cells.
Figure 5.
DSBs at the nuclear lamina are positionally stable. (A) Immunofluorescence of HT1080 cells expressing Dam-LaminB1 and m6A-Tracer 2 h after treatment (or not) with 50 ng/mL neocarzinostatin (NCS) for 15 min. (B) Box plot of GFP intensity ratios of the signal in the nucleoplasm versus the signal at the nuclear envelope in a HT1080-derived clonal cell line expressing a Dam-LaminB1 and the m6A-Tracer. The number of cells analyzed per condition was 20. For statistical analysis, χ2 tests were performed. (n.s.) Nonsignificant. (C) dSTORM microscopy images of LADs (green) and laminB (left panel; red) or TPR (right panel; red) in the absence (top panel) or presence (bottom panel) of DNA damage (100 ng/mL NCS for 15 min and released for 2 h) in HT1080 cells expressing Dam-LaminB1 and m6A-Tracer. Images were taken from the bottom of the cells to allow better resolution of nuclear pores. Corresponding colocalization and the ratio of positive over negative colocalization events are displayed at the right. The mean ratios for all nuclei analyzed (n ≥ 8) are displayed above.
Figure 6.
DSBs at the nuclear lamina are repaired by NHEJ or A-EJ. The percentage of colocalization of the lacO array with γ-H2AX in untreated cells (NT) or after 14 h of Dox (time point 0) and subsequent release for 24 h in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD and transfected with XRCC4 (A), RAD51 (B), ligase 3 (C), XRCC1 (D), or PARP1-specific siRNAs (E) is shown. (F) The percentage of colocalization of the lacO array with γ-H2AX upon Dox treatment or release in I-U2OS19 cells expressing GFP-lacI or GFP-lacI-ΔEMD and treated with DMSO or a PARP inhibitor (PARPi, during the entire course of the experiment) is shown. Values represent mean ± SD of three independent experiments with n > 50 cells. For statistical analysis, a _t_-test was performed. (*) P < 0.05; (**) P < 0.01.
Similar articles
- DSB (Im)mobility and DNA repair compartmentalization in mammalian cells.
Lemaître C, Soutoglou E. Lemaître C, et al. J Mol Biol. 2015 Feb 13;427(3):652-8. doi: 10.1016/j.jmb.2014.11.014. Epub 2014 Nov 24. J Mol Biol. 2015. PMID: 25463437 Review. - DNA double strand break repair pathway choice: a chromatin based decision?
Clouaire T, Legube G. Clouaire T, et al. Nucleus. 2015;6(2):107-13. doi: 10.1080/19491034.2015.1010946. Epub 2015 Feb 12. Nucleus. 2015. PMID: 25675367 Free PMC article. - Double strand break (DSB) repair in heterochromatin and heterochromatin proteins in DSB repair.
Lemaître C, Soutoglou E. Lemaître C, et al. DNA Repair (Amst). 2014 Jul;19:163-8. doi: 10.1016/j.dnarep.2014.03.015. Epub 2014 Apr 19. DNA Repair (Amst). 2014. PMID: 24754998 Review. - Inhibition of homologous recombination by hyperthermia shunts early double strand break repair to non-homologous end-joining.
Bergs JW, Krawczyk PM, Borovski T, ten Cate R, Rodermond HM, Stap J, Medema JP, Haveman J, Essers J, van Bree C, Stalpers LJ, Kanaar R, Aten JA, Franken NA. Bergs JW, et al. DNA Repair (Amst). 2013 Jan 1;12(1):38-45. doi: 10.1016/j.dnarep.2012.10.008. Epub 2012 Dec 11. DNA Repair (Amst). 2013. PMID: 23237939 - Homologous recombination protects mammalian cells from replication-associated DNA double-strand breaks arising in response to methyl methanesulfonate.
Nikolova T, Ensminger M, Löbrich M, Kaina B. Nikolova T, et al. DNA Repair (Amst). 2010 Oct 5;9(10):1050-63. doi: 10.1016/j.dnarep.2010.07.005. Epub 2010 Aug 13. DNA Repair (Amst). 2010. PMID: 20708982
Cited by
- The Determinant of DNA Repair Pathway Choices in Ionising Radiation-Induced DNA Double-Strand Breaks.
Zhao L, Bao C, Shang Y, He X, Ma C, Lei X, Mi D, Sun Y. Zhao L, et al. Biomed Res Int. 2020 Aug 25;2020:4834965. doi: 10.1155/2020/4834965. eCollection 2020. Biomed Res Int. 2020. PMID: 32908893 Free PMC article. Review. - Complex Chromatin Motions for DNA Repair.
Miné-Hattab J, Chiolo I. Miné-Hattab J, et al. Front Genet. 2020 Aug 27;11:800. doi: 10.3389/fgene.2020.00800. eCollection 2020. Front Genet. 2020. PMID: 33061931 Free PMC article. Review. - Regulation of Nuclear Mechanics and the Impact on DNA Damage.
Dos Santos Á, Toseland CP. Dos Santos Á, et al. Int J Mol Sci. 2021 Mar 20;22(6):3178. doi: 10.3390/ijms22063178. Int J Mol Sci. 2021. PMID: 33804722 Free PMC article. Review. - Efficient Pre-mRNA Cleavage Prevents Replication-Stress-Associated Genome Instability.
Teloni F, Michelena J, Lezaja A, Kilic S, Ambrosi C, Menon S, Dobrovolna J, Imhof R, Janscak P, Baubec T, Altmeyer M. Teloni F, et al. Mol Cell. 2019 Feb 21;73(4):670-683.e12. doi: 10.1016/j.molcel.2018.11.036. Epub 2019 Jan 10. Mol Cell. 2019. PMID: 30639241 Free PMC article. - Structural variations in cancer and the 3D genome.
Dubois F, Sidiropoulos N, Weischenfeldt J, Beroukhim R. Dubois F, et al. Nat Rev Cancer. 2022 Sep;22(9):533-546. doi: 10.1038/s41568-022-00488-9. Epub 2022 Jun 28. Nat Rev Cancer. 2022. PMID: 35764888 Free PMC article. Review.
References
- Bickmore WA. 2013. The spatial organization of the human genome. Annu Rev Genomics Hum Genet 14: 67–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources