Spatial localization of recent ancestors for admixed individuals - PubMed (original) (raw)
Spatial localization of recent ancestors for admixed individuals
Wen-Yun Yang et al. G3 (Bethesda). 2014.
Abstract
Ancestry analysis from genetic data plays a critical role in studies of human disease and evolution. Recent work has introduced explicit models for the geographic distribution of genetic variation and has shown that such explicit models yield superior accuracy in ancestry inference over nonmodel-based methods. Here we extend such work to introduce a method that models admixture between ancestors from multiple sources across a geographic continuum. We devise efficient algorithms based on hidden Markov models to localize on a map the recent ancestors (e.g., grandparents) of admixed individuals, joint with assigning ancestry at each locus in the genome. We validate our methods by using empirical data from individuals with mixed European ancestry from the Population Reference Sample study and show that our approach is able to localize their recent ancestors within an average of 470 km of the reported locations of their grandparents. Furthermore, simulations from real Population Reference Sample genotype data show that our method attains high accuracy in localizing recent ancestors of admixed individuals in Europe (an average of 550 km from their true location for localization of two ancestries in Europe, four generations ago). We explore the limits of ancestry localization under our approach and find that performance decreases as the number of distinct ancestries and generations since admixture increases. Finally, we build a map of expected localization accuracy across admixed individuals according to the location of origin within Europe of their ancestors.
Keywords: admixture; ancestry inference; genetic continuum; genetic variation; localization.
Copyright © 2014 Yang et al.
Figures
Figure 1
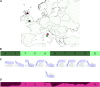
SPAMIX model for admixed individuals. (A) Example of haploid individual with two ancestry locations in Europe (circles denote the true ancestry locations). (B) The admixture process induces segments of different ancestry backgrounds. (C) SPAMIX uses logistic gradients to describe allele frequencies as a function of geographic map to instantiate an admixture hidden Markov modeling for each pair of locations on a map. Each location on the map is associated to a particular allele frequency at all sites in the genome. (D) SPAMIX finds the location of ancestors on a map (denoted by squares in A) and the locus-specific ancestry at each site in the genome by maximizing the likelihood of genotype data.
Figure 2
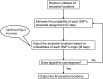
An illustration of the expectation-maximization (EM) algorithm for spatial ancestry inference for haploid data. The E-step and M-step are performed alternatively until the EM algorithm converges. The last M ancestral locations are used as the output of EM algorithm. SNPs, single-nucleotide polymorphisms.
Figure 3
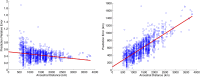
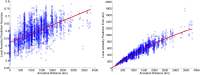
Ancestral location prediction error as a function of distance between ancestral locations in simulations over Population Reference Sample data. Left, the prediction error normalized by the distance between the ancestral locations used in simulations; right, plot of the prediction error. Simulations use the haploid model with two generations in the mixture.
Figure 4
Inference of number of distinct ancestries using the Akaike information criterion (AIC). We simulated 1000 admixed individuals with up to four distinct ancestry sources in Europe and used the AIC within the SPAMIX model to infer the number of ancestries. (A−D) Proportion of inferred number of ancestries (y-axis) as function of number of simulated ancestries (x-axis). Although we observed a large variance in the number of predicted ancestries, we note that the histogram is centered on the correct simulated number of ancestries, thus suggesting that AIC could be used to infer the number of distinct ancestors.
Figure 5
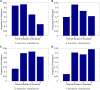
SPAMIX locus-specific ancestry prediction accuracy as function of distance between ancestral locations. Left, local ancestry prediction accuracy, defined as the percentage of all loci with correct assignment of ancestry. Right, average distance to true locations for each allele in the genome (local ancestry prediction error). Simulations use the haploid model with two generations in the mixture.
Figure 6
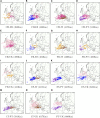
Ancestral location prediction error in simulations of European individuals with ancestry from two locations in Europe, stratified by the country of origin of each location (the country of origin is displayed in different colors). The assumed true locations are displayed by shaded circles. Results in parenthesis denote the average ancestral location prediction error across all simulations. In each simulation the reference data (used to estimate logistic gradients) is disjoint from data used to simulate admixed genomes (see the section Materials and Methods). The admixed genome is simulated as four generations ago, and SPAMIX diploid model is used for the inference. The number of simulated pairs can be found in
Figure S3
.
Figure 7
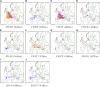
Ancestral location prediction error in real POPRES admixed individuals, stratified by the country of origin of each location. Letters are the inferred locations, and the shaded circles are the assumed true locations.
Similar articles
- Characterizing features affecting local ancestry inference performance in admixed populations.
Honorato-Mauer J, Shah NN, Maihofer AX, Zai CC, Belangero S, Nievergelt CM; Psychiatric Genomics Consortium for PTSD Ancestry Working Group; Santoro M, Atkinson EG. Honorato-Mauer J, et al. Am J Hum Genet. 2025 Feb 6;112(2):224-234. doi: 10.1016/j.ajhg.2024.12.005. Epub 2025 Jan 2. Am J Hum Genet. 2025. PMID: 39753130 - The Analysis of Ethnic Mixtures.
Zhu X, Wang H. Zhu X, et al. Methods Mol Biol. 2017;1666:505-525. doi: 10.1007/978-1-4939-7274-6_25. Methods Mol Biol. 2017. PMID: 28980262 - A spatial haplotype copying model with applications to genotype imputation.
Yang WY, Hormozdiari F, Eskin E, Pasaniuc B. Yang WY, et al. J Comput Biol. 2015 May;22(5):451-62. doi: 10.1089/cmb.2014.0151. Epub 2014 Dec 19. J Comput Biol. 2015. PMID: 25526526 Free PMC article. - Human genetic admixture.
Korunes KL, Goldberg A. Korunes KL, et al. PLoS Genet. 2021 Mar 11;17(3):e1009374. doi: 10.1371/journal.pgen.1009374. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33705374 Free PMC article. Review. - Analyses of genetic ancestry enable key insights for molecular ecology.
Gompert Z, Buerkle CA. Gompert Z, et al. Mol Ecol. 2013 Nov;22(21):5278-94. doi: 10.1111/mec.12488. Epub 2013 Sep 19. Mol Ecol. 2013. PMID: 24103088 Review.
Cited by
- Toward high-resolution population genomics using archaeological samples.
Morozova I, Flegontov P, Mikheyev AS, Bruskin S, Asgharian H, Ponomarenko P, Klyuchnikov V, ArunKumar G, Prokhortchouk E, Gankin Y, Rogaev E, Nikolsky Y, Baranova A, Elhaik E, Tatarinova TV. Morozova I, et al. DNA Res. 2016 Aug;23(4):295-310. doi: 10.1093/dnares/dsw029. Epub 2016 Jul 19. DNA Res. 2016. PMID: 27436340 Free PMC article. Review. - Application of the geographic population structure (GPS) algorithm for biogeographical analyses of wild and captive gorillas.
Das R, Upadhyai P. Das R, et al. BMC Bioinformatics. 2019 Feb 5;20(Suppl 1):35. doi: 10.1186/s12859-018-2568-5. BMC Bioinformatics. 2019. PMID: 30717677 Free PMC article. - Differential Evolution approach to detect recent admixture.
Kozlov K, Chebotarev D, Hassan M, Triska M, Triska P, Flegontov P, Tatarinova TV. Kozlov K, et al. BMC Genomics. 2015;16 Suppl 8(Suppl 8):S9. doi: 10.1186/1471-2164-16-S8-S9. Epub 2015 Jun 18. BMC Genomics. 2015. PMID: 26111206 Free PMC article. - Inferring the ancestry of parents and grandparents from genetic data.
Pei J, Zhang Y, Nielsen R, Wu Y. Pei J, et al. PLoS Comput Biol. 2020 Aug 14;16(8):e1008065. doi: 10.1371/journal.pcbi.1008065. eCollection 2020 Aug. PLoS Comput Biol. 2020. PMID: 32797037 Free PMC article.
References
- Bozdogan H., 1987. Model selection and Akaike’s information criterion (AIC): The general theory and its analytical extensions. Psychometrika 52: 345–370.
Publication types
MeSH terms
Grants and funding
- R03 CA162200/CA/NCI NIH HHS/United States
- R01 HG007089/HG/NHGRI NIH HHS/United States
- R01 GM053275/GM/NIGMS NIH HHS/United States
- R01-GM053275/GM/NIGMS NIH HHS/United States
- R03-CA162200/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources