Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein - PubMed (original) (raw)
. 2014 Oct 23;9(2):495-503.
doi: 10.1016/j.celrep.2014.09.036. Epub 2014 Oct 16.
Jiayi Yang 2, Che-Hung Shen 1, Eric C Peters 2, Julien Fitamant 3, Puiyee Chan 1, Mindy Hsieh 2, Shunying Zhu 1, John M Asara 4, Bin Zheng 1, Nabeel Bardeesy 3, Jun Liu 2, Xu Wu 5
Affiliations
- PMID: 25373897
- PMCID: PMC4223634
- DOI: 10.1016/j.celrep.2014.09.036
Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein
Michael DeRan et al. Cell Rep. 2014.
Abstract
Hippo signaling is a tumor-suppressor pathway involved in organ size control and tumorigenesis through the inhibition of YAP and TAZ. Here, we show that energy stress induces YAP cytoplasmic retention and S127 phosphorylation and inhibits YAP transcriptional activity and YAP-dependent transformation. These effects require the central metabolic sensor AMP-activated protein kinase (AMPK) and the upstream Hippo pathway components Lats1/Lats2 and angiomotin-like 1 (AMOTL1). Furthermore, we show that AMPK directly phosphorylates S793 of AMOTL1. AMPK activation stabilizes and increases AMOTL1 steady-state protein levels, contributing to YAP inhibition. The phosphorylation-deficient S793Ala mutant of AMOTL1 showed a shorter half-life and conferred resistance to energy-stress-induced YAP inhibition. Our findings link energy sensing to the Hippo-YAP pathway and suggest that YAP may integrate spatial (contact inhibition), mechanical, and metabolic signals to control cellular proliferation and survival.
Figures
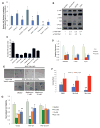
Figure 1. Energy stress inhibits YAP
A, Confluent and serum stimulated HEK293A cells were treated with DMSO control, AICAR (1mM), metformin (10mM) or phenformin (1mM), with or without Compound C (10 μM) for 3h. YAP nuclear localization is quantified by determining the Pearson’s correlation coefficient of nuclear staining. Data are represented as mean, S.D. n=3. (*, p<0.03; **, p<0.001). B, Small molecule energy stressors induce YAP S127 and TAZ Ser89 phosphorylation in HaCaT cells. HaCaT cells were treated with DMSO (control), 1 mM phenformin, 1 mM AICAR or 25 mM 2-DG for 6h. C, Inhibition of YAP-reporter activity. HEK293 YAP reporter cells were treated with DMSO control, 2-DG (8.3 or 25 mM), AICAR (1 or 3 mM) or Metformin (10 mM). (data are represented as mean, S.D. n=3). D, Energy stressors suppress YAP target gene expression. HEK293A cells were treated with DMSO (ctrl), 1mM phenformin, or 1mM AICAR for 16 hours. Relative mRNA levels are normalized to DMSO (ctrl). E, MCF10A cells transduced with a vector control, YAP wild type or YAP S127A mutant were grown on Matrigel without EGF. Cells were treated with metformin (2mM or 10mM) or DMSO control. Scale bar: 100 μm. F, The colony numbers per field are quantified by Image J, and shown in bar graph (data are represented as mean, SD, n=4; *, p<0.05; **, p<0.0005 by comparing to the S127A mutant samples with the same treatment). G, Metformin dose dependently inhibits the proliferation of primary mouse hepatocellular carcinoma (HCC) cell line JF001 (_Mst1_−/−_Mst2_−/−). JF001 cells transduced with vector, wild-type YAP, or YAP S127A mutant were treated as indicated. (Data are represented as mean, SD, n=3; *, p<0.05; **, p<0.001 by comparing to the DMSO control). See also Figure S1.
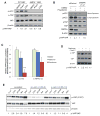
Figure 2. Energy stress inhibits YAP through AMP-activated protein kinase (AMPK) and Lats1/2
A, AMPK is required for YAP inhibition. Wild type or AMPK−/− (AMPKα1α2 double knockout) MEFs were treated with DMSO control, 2-DG (25 mM) or phenformin (1 mM) for 12h. Cells were then harvested for western blot analysis. B, HaCaT cells were transfected with siRNAs targeting AMPKα1 and α2, and then treated for 16h with DMSO or phenformin (1mM). C, siRNAs targeting AMPKα1 and α2 blocks YAP cytoplasmic retention. Cells were transfected with siRNAs targeting AMPK, and then treated for 16h with DMSO, metformin (10mM) or phenformin (1mM). YAP nuclear localization is quantified by determining the Pearson’s correlation coefficient between YAP positive areas with the nuclear staining. Data are represented as mean, S.D. n=3. (*, p<0.05; **, p<0.001 by comparing to the DMSO control). D, HCCs were isolated from tumors derived from liver specific _Mst1_−/−_Mst2_−/− mice. Cells are treated with metformin (10mM) or co-treated with Compound C (10 μM) for 8h. p-YAP levels were evaluated by western blot. E, HEK293A cells were transfected with siRNA control or siRNA targeting Lats1 and Lats2 (#5+8, or #5+10). Cells were treated with AICAR (1mM) or Metformin (10mM) for 8h, serum free media (SFM, 3h) or 2-DG (25mM, 3h). Cells were then harvested for western blot analysis of p-YAP (Ser127). See also Figure S2.
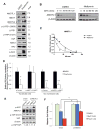
Figure 3. Endogenous angiomotin like-1 (AMOTL1) is stabilized in response to energy stress and is required to mediate AMPK-induced YAP-cytoplasmic retention and Ser127 phosphorylation
A, HEK293A cells were treated with DMSO control or phenformin (1 mM) for 16h. Protein levels of Hippo pathway components (AMOTL1, AMOT, AMOTL2, p-YAP, YAP, p-Lats1, Lats1, MST2, and NF2) were analyzed by western blots. B, Cells were treated with metformin (10mM) for 4h and with cycloheximide (CHX) with indicated time. Protein levels of endogenous AMOTL1 were analyzed by western blots. C, Protein levels from B were quantified by densitometry of the bands. Projected degradation curve of AMOTL1 was plotted and fitted using Prism software. D, HEK293A cells were transfected with siRNAs targeting AMOT and AMOTL1, with GFP to mark transfected cells. Cells were treated with phenformin (1mM) or DMSO. YAP nuclear localization is quantified by determining the Pearson’s correlation coefficient, in GFP positive (transfected) or GFP negative (non-transfected) cells. Data are represented as mean, S.D. n=3. (*, p<0.02; **, p<0.001 by comparing to the DMSO controls). E. HaCaT cells were transfected with shRNA targeting AMOTL1. Cells were treated with DMSO control or phenformin (1mM) and p-YAP level is analyzed. F. Cells were transfected shRNA targeting AMOTL1, and then treated with DMSO, metformin (10mM) or phenformin (1mM) for 48 hours. The cell proliferation is determined by measuring the cell viability (*, p<0.01). See also Figure S3.
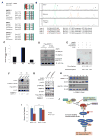
Figure 4. AMPK directly phosphorylates AMOTL1 Ser793 leading to its stabilization and YAP inhibition
A, Alignment of the conserved AMPK substrate motifs in angiomotin family proteins (AMOTL1, AMOT and AMOTL2) of different species. B, LC-MS/MS studies of phosphorylation of AMOTL1 Ser793. Flag-AMOTL1 was purified from HEK293A cells treated with metformin (10mM) or metformin and C ompound C (10μM). Extracted ion chromatographs of peptides containing Ser793 were shown. Peak A (unphosphoryated peptide), Peak B (peptide with p-Ser805), Peak C (peptide with p-Ser793), Peak D (peptide with p-Ser793 and p-Ser805). C, The abundance of the peptides was quantified by measuring the area under the curve (AUC) in the spectra. D, Recombinant AMPK phosphorylates AMOTL1 wild type, but not S793A mutant in vitro. E, HEK239T cells were treated for 8 hours with DMSO (control), 1μM A-769662, or 1mM Phenformin. Protein lysates were then analyzed by Phos-Tag SDS-PAGE followed by Western blotting with an anti-AMOTL1 antibody. F, Phenformin treatment increased AMOTL1 S793 phosphorylation. An antibody recognizing p-AMOTL1 (Ser793) was used in western blot. HaCaT cells were transfected with Flag-AMOTL1, and then treated with DMSO, or phenformin (1mM) for 1hr or 2hr. G, Knock-down of AMPK decreased the levels of p-AMOTL1. Cells were transfected with siRNAs targeting AMPKα1 and α2, and Flag-AMOTL1. Cells were then treated with metformin (10mM) or DMSO control. p-AMOTL1 levels were determined by using a phosphospecific antibody recognizing AMOTL1 Ser793. H, AMOTL1 S793A has a shorter half-life than wild type AMOTL1. HEK293T cells transfected with either wild type or S793A mutant AMOTL1 were treated for 16 hours with A-769662 then treated for the indicated time with cycloheximide (CHX). I. Cells were transfected with AMOTL1 wild type or S793A mutant, and then treated with DMSO control, or phenformin (1mM) for 48 hours. The cell viability is determined by CellTiter Glo (*, p<0.01). J. A proposed model of energy stress-mediated YAP inhibition. See also Figure S4.
Similar articles
- Angiomotins stimulate LATS kinase autophosphorylation and act as scaffolds that promote Hippo signaling.
Mana-Capelli S, McCollum D. Mana-Capelli S, et al. J Biol Chem. 2018 Nov 23;293(47):18230-18241. doi: 10.1074/jbc.RA118.004187. Epub 2018 Sep 28. J Biol Chem. 2018. PMID: 30266805 Free PMC article. - Sphingosylphosphorylcholine regulates the Hippo signaling pathway in a dual manner.
Kemppainen K, Wentus N, Lassila T, Laiho A, Törnquist K. Kemppainen K, et al. Cell Signal. 2016 Dec;28(12):1894-1903. doi: 10.1016/j.cellsig.2016.09.004. Epub 2016 Sep 12. Cell Signal. 2016. PMID: 27634386 - Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases.
Adler JJ, Johnson DE, Heller BL, Bringman LR, Ranahan WP, Conwell MD, Sun Y, Hudmon A, Wells CD. Adler JJ, et al. Proc Natl Acad Sci U S A. 2013 Oct 22;110(43):17368-73. doi: 10.1073/pnas.1308236110. Epub 2013 Oct 7. Proc Natl Acad Sci U S A. 2013. PMID: 24101513 Free PMC article. - Angiomotin'g YAP into the nucleus for cell proliferation and cancer development.
Hong W. Hong W. Sci Signal. 2013 Sep 3;6(291):pe27. doi: 10.1126/scisignal.2004573. Sci Signal. 2013. PMID: 24003252 Review. - The Hippo pathway in normal development and cancer.
Maugeri-Saccà M, De Maria R. Maugeri-Saccà M, et al. Pharmacol Ther. 2018 Jun;186:60-72. doi: 10.1016/j.pharmthera.2017.12.011. Epub 2018 Jan 3. Pharmacol Ther. 2018. PMID: 29305295 Review.
Cited by
- YAP Inhibition by Resveratrol via Activation of AMPK Enhances the Sensitivity of Pancreatic Cancer Cells to Gemcitabine.
Jiang Z, Chen X, Chen K, Sun L, Gao L, Zhou C, Lei M, Duan W, Wang Z, Ma Q, Ma J. Jiang Z, et al. Nutrients. 2016 Sep 23;8(10):546. doi: 10.3390/nu8100546. Nutrients. 2016. PMID: 27669292 Free PMC article. - Hippo signalling during development.
Davis JR, Tapon N. Davis JR, et al. Development. 2019 Sep 16;146(18):dev167106. doi: 10.1242/dev.167106. Development. 2019. PMID: 31527062 Free PMC article. Review. - The Hippo Signaling Pathway in Development and Disease.
Zheng Y, Pan D. Zheng Y, et al. Dev Cell. 2019 Aug 5;50(3):264-282. doi: 10.1016/j.devcel.2019.06.003. Dev Cell. 2019. PMID: 31386861 Free PMC article. Review. - Insulin suppresses transcriptional activity of yes-associated protein in insulin target cells.
Sayedyahossein S, Hedman AC, Sacks DB. Sayedyahossein S, et al. Mol Biol Cell. 2020 Jan 15;31(2):131-141. doi: 10.1091/mbc.E19-04-0205. Epub 2019 Nov 6. Mol Biol Cell. 2020. PMID: 31693448 Free PMC article. - Metformin accelerates bone fracture healing by promoting type H vessel formation through inhibition of YAP1/TAZ expression.
Ruan Z, Yin H, Wan TF, Lin ZR, Zhao SS, Long HT, Long C, Li ZH, Liu YQ, Luo H, Cheng L, Chen C, Zeng M, Lin ZY, Zhao RB, Chen CY, Wang ZX, Liu ZZ, Cao J, Wang YY, Jin L, Liu YW, Zhu GQ, Zou JT, Gong JS, Luo Y, Hu Y, Zhu Y, Xie H. Ruan Z, et al. Bone Res. 2023 Aug 16;11(1):45. doi: 10.1038/s41413-023-00279-4. Bone Res. 2023. PMID: 37587136 Free PMC article.
References
- Adler JJ, Johnson DE, Heller BL, Bringman LR, Ranahan WP, Conwell MD, Sun Y, Hudmon A, Wells CD. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci U S A. 2013b;110:17368–17373. - PMC - PubMed
- Cool B, Zinker B, Chiou W, Kifle L, Cao N, Perham M, Dickinson R, Adler A, Gagne G, Iyengar R, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3:403–416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA120964/CA/NCI NIH HHS/United States
- R01 CA166717/CA/NCI NIH HHS/United States
- R01CA181537/CA/NCI NIH HHS/United States
- R01 CA181537/CA/NCI NIH HHS/United States
- R01CA166717/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases