TCR triggering by pMHC ligands tethered on surfaces via poly(ethylene glycol) depends on polymer length - PubMed (original) (raw)
TCR triggering by pMHC ligands tethered on surfaces via poly(ethylene glycol) depends on polymer length
Zhengyu Ma et al. PLoS One. 2014.
Abstract
Antigen recognition by T cells relies on the interaction between T cell receptor (TCR) and peptide-major histocompatibility complex (pMHC) at the interface between the T cell and the antigen presenting cell (APC). The pMHC-TCR interaction is two-dimensional (2D), in that both the ligand and receptor are membrane-anchored and their movement is limited to 2D diffusion. The 2D nature of the interaction is critical for the ability of pMHC ligands to trigger TCR. The exact properties of the 2D pMHC-TCR interaction that enable TCR triggering, however, are not fully understood. Here, we altered the 2D pMHC-TCR interaction by tethering pMHC ligands to a rigid plastic surface with flexible poly(ethylene glycol) (PEG) polymers of different lengths, thereby gradually increasing the ligands' range of motion in the third dimension. We found that pMHC ligands tethered by PEG linkers with long contour length were capable of activating T cells. Shorter PEG linkers, however, triggered TCR more efficiently. Molecular dynamics simulation suggested that shorter PEGs exhibit faster TCR binding on-rates and off-rates. Our findings indicate that TCR signaling can be triggered by surface-tethered pMHC ligands within a defined 3D range of motion, and that fast binding rates lead to higher TCR triggering efficiency. These observations are consistent with a model of TCR triggering that incorporates the dynamic interaction between T cell and antigen-presenting cell.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
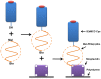
Figure 1. Schematic illustration of IEkMCC ligands tethered onto a plastic surface with PEG polymer linkers.
IEkMCC proteins with free c-terminal cysteines were first conjugated with heterobifunctional PEG linkers Mal-PEG-Bio through interactions between the sulfhydryl group and the maleimide group. Conjugates with biotin at the free ends of the polymer were then tethered to a plastic surface coated with streptavidin.
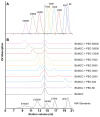
Figure 2. Characterization and separation of PEG polymer linkers and IEkMCC-PEG conjugates.
(A) Compiled elution curves of nine PEG polymer linkers from a Superdex 200 10/300 GL gel filtration column. The polymers were detected through the weak UV absorption of the biotin group using a 245 nm UV detector. (B) Separation of the IEkMCC and PEG polymer reaction products. The reaction products were loaded on a Superdex 200 10/300 GL gel filtration column to separate IEkMCC-PEG conjugates, unreacted IEkMCC, and unreacted PEG polymers. The reaction products of PEG 15000, PEG 30000 and PEG 60000 were first purified with an IEk-binding affinity column to eliminate unreacted PEG polymers. The dotted vertical line indicates the elution volume of IEkMCC protein. The late elution peaks of unreacted polymers can be seen for PEGs ranging from PEG 88 to PEG 5000. In reaction with PEG 7500, unreacted IEkMCC and unreacted polymer formed a single peak that was eluted at a position between unconjugated IEkMCC and pure PEG 7500.
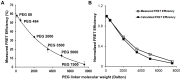
Figure 3. FRET between streptavidin on plastic plates and IEkMCC tethered with PEG polymers.
(A) Measured FRET efficiencies of IEkMCC tethered with six different PEG polymers. The intensity of DyLight 549 was captured before and after DyLight 649 was photobleached. The measured FRET efficiency ( ) was calculated using the intensity of DyLight 549 before (
) was calculated using the intensity of DyLight 549 before ( ) and after (
) and after ( ) DyLight 649 photobleaching (
) DyLight 649 photobleaching ( ). The averaged values of two measurements were plotted with standard deviations. (B) After normalization, the measured FRET efficiencies match those calculated based on the Flory radius (
). The averaged values of two measurements were plotted with standard deviations. (B) After normalization, the measured FRET efficiencies match those calculated based on the Flory radius ( ) of the PEG polymers. The
) of the PEG polymers. The  of the PEG polymer of
of the PEG polymer of  subunits and unit length
subunits and unit length  was calculated using
was calculated using  , where
, where  is 0.28 nm . Theoretical FRET efficiency (
is 0.28 nm . Theoretical FRET efficiency ( ) was calculated using the equation
) was calculated using the equation  , where the Förster distance (
, where the Förster distance ( ) of the DyLight 549-DyLight 649 donor-acceptor pair is 5 nm and the distance between the pMHC ligand and streptavidin
) of the DyLight 549-DyLight 649 donor-acceptor pair is 5 nm and the distance between the pMHC ligand and streptavidin  is
is  of the PEG polymer plus the pMHC radius of 2 nm. The FRET efficiencies were normalized by dividing the FRET efficiencies by the FRET efficiency of PEG 88.
of the PEG polymer plus the pMHC radius of 2 nm. The FRET efficiencies were normalized by dividing the FRET efficiencies by the FRET efficiency of PEG 88.
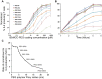
Figure 4. T cell activation by IEkMCC tethered with PEG polymers of different lengths.
(A) T cell IL2 production in response to IEkMCC-PEG ligands of varying coating densities after 6 hours of stimulation. Data are representative of three independent experiments. The percent of T cells producing IL2 was determined by intracellular staining and flow cytometry. Three experiments using T cells from three different mice were performed (see Fig. S8 for flow cytometry plots). The percent of T cells producing IL2 was normalized to the highest value in each experiment. The data points are averages of the normalized values with standard errors of the means. (B) The rate of T cell response to IEkMCC ligands tethered with PEG polymers of different lengths. T cell IL2 production in response to stimulation on 96 well plates coated with 110 pM IEkMC-PEG ligands. T cells were harvested every hour for 6 hours and levels of IL2 expression were assayed by flow cytometry. Three experiments using T cells from three different mice were performed (see Fig. S9 for flow cytometry plots). The percent of T cells producing IL2 was normalized to the highest value in each experiment. The data points are averages of the normalized values with standard errors of the means. (C) The rates of T cell IL2 responses to IEkMCC ligands tethered with PEG polymers were extracted from the slope of linear fitting curves in Fig. 4B and plotted against the Flory radius of the polymers. The linear regressions and equations for deriving the rates are shown in Fig. S7.
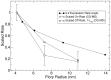
Figure 5. The impact of polymer length on pMHC-TCR binding kinetics based on CG-MD simulation.
The on-rates and off-rates derived from the simulation for the PEG 4000, PEG 10000 and PEG 20000 were scaled against the data for the PEG 4000 and plotted as a function of the polymer Flory radius. To display the relationship between binding rates and TCR triggering efficiency, the experimentally determined IL2 expression rates for PEG 3500, PEG 5000, PEG 7500, PEG 150000 and PEG 30000 shown in Fig. 4A were scaled against PEG 3500 and plotted as a function of the polymer Flory radius.
Similar articles
- T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand.
Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. Choudhuri K, et al. Nature. 2005 Jul 28;436(7050):578-82. doi: 10.1038/nature03843. Nature. 2005. PMID: 16049493 - Modulation of T cell function by TCR/pMHC binding kinetics.
Carreño LJ, González PA, Kalergis AM. Carreño LJ, et al. Immunobiology. 2006;211(1-2):47-64. doi: 10.1016/j.imbio.2005.09.003. Epub 2006 Jan 4. Immunobiology. 2006. PMID: 16446170 Review. - The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness.
Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. Huang J, et al. Nature. 2010 Apr 8;464(7290):932-6. doi: 10.1038/nature08944. Epub 2010 Mar 31. Nature. 2010. PMID: 20357766 Free PMC article. - A flexible docking approach for prediction of T cell receptor-peptide-MHC complexes.
Pierce BG, Weng Z. Pierce BG, et al. Protein Sci. 2013 Jan;22(1):35-46. doi: 10.1002/pro.2181. Protein Sci. 2013. PMID: 23109003 Free PMC article. - Insights from in situ analysis of TCR-pMHC recognition: response of an interaction network.
Zhu C, Jiang N, Huang J, Zarnitsyna VI, Evavold BD. Zhu C, et al. Immunol Rev. 2013 Jan;251(1):49-64. doi: 10.1111/imr.12016. Immunol Rev. 2013. PMID: 23278740 Free PMC article. Review.
Cited by
- Hierarchically Multivalent Peptide-Nanoparticle Architectures: A Systematic Approach to Engineer Surface Adhesion.
Jeong WJ, Bu J, Jafari R, Rehak P, Kubiatowicz LJ, Drelich AJ, Owen RH, Nair A, Rawding PA, Poellmann MJ, Hopkins CM, Král P, Hong S. Jeong WJ, et al. Adv Sci (Weinh). 2022 Feb;9(4):e2103098. doi: 10.1002/advs.202103098. Epub 2021 Dec 11. Adv Sci (Weinh). 2022. PMID: 34894089 Free PMC article. - Targeting CD4+ Cells with Anti-CD4 Conjugated Mertansine-Loaded Nanogels.
Canakci M, Singh K, Munkhbat O, Shanthalingam S, Mitra A, Gordon M, Osborne BA, Thayumanavan S. Canakci M, et al. Biomacromolecules. 2020 Jun 8;21(6):2473-2481. doi: 10.1021/acs.biomac.0c00442. Epub 2020 May 28. Biomacromolecules. 2020. PMID: 32383874 Free PMC article. - Quantifying the distinct role of plasmon enhancement mechanisms in prototypical antenna-reactor photocatalysts.
Liu S, Wu Z, Zhu Z, Feng K, Zhou Y, Hu X, Huang X, Zhang B, Dong X, Ma Y, Nie K, Shen J, Wang Z, He J, Wang J, Ji Y, Yan B, Zhang Q, Genest A, Zhang X, Li C, Wu B, An X, Rupprechter G, He L. Liu S, et al. Nat Commun. 2025 Mar 6;16(1):2245. doi: 10.1038/s41467-025-57569-0. Nat Commun. 2025. PMID: 40050268 Free PMC article. - Single electron transfer events and dynamical heterogeneity in the small protein azurin from Pseudomonas aeruginosa.
Pradhan B, Engelhard C, Van Mulken S, Miao X, Canters GW, Orrit M. Pradhan B, et al. Chem Sci. 2019 Nov 27;11(3):763-771. doi: 10.1039/c9sc05405g. Chem Sci. 2019. PMID: 34123050 Free PMC article. - Robust membrane protein tweezers reveal the folding speed limit of helical membrane proteins.
Kim S, Lee D, Wijesinghe WCB, Min D. Kim S, et al. Elife. 2023 May 30;12:e85882. doi: 10.7554/eLife.85882. Elife. 2023. PMID: 37249211 Free PMC article.
References
- van der Merwe PA (2001) The TCR triggering puzzle. Immunity 14: 665–668. - PubMed
- van der Merwe PA, Dushek O (2011) Mechanisms for T cell receptor triggering. Nat Rev Immunol 11: 47–55. - PubMed
- Davis SJ, van der Merwe PA (2006) The kinetic-segregation model: TCR triggering and beyond. Nat Immunol 7: 803–809. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 GM103464/GM/NIGMS NIH HHS/United States
- R21 AI078387/AI/NIAID NIH HHS/United States
- R21 AI087516/AI/NIAID NIH HHS/United States
- 2-P30-AI-045008-11/AI/NIAID NIH HHS/United States
- 1R21 AI078387/AI/NIAID NIH HHS/United States
- P20GM103464/GM/NIGMS NIH HHS/United States
- 1R21 AI087516/AI/NIAID NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- F32 CA134157/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources