A reference bacterial genome dataset generated on the MinION™ portable single-molecule nanopore sequencer - PubMed (original) (raw)
A reference bacterial genome dataset generated on the MinION™ portable single-molecule nanopore sequencer
Joshua Quick et al. Gigascience. 2014.
Erratum in
- Erratum: A reference bacterial genome dataset generated on the MinION(TM) portable single-molecule nanopore sequencer.
Quick J, Quinlan AR, Loman NJ. Quick J, et al. Gigascience. 2015 Feb 13;4:6. doi: 10.1186/s13742-015-0043-z. eCollection 2015. Gigascience. 2015. PMID: 25695306 Free PMC article.
Abstract
Background: The MinION™ is a new, portable single-molecule sequencer developed by Oxford Nanopore Technologies. It measures four inches in length and is powered from the USB 3.0 port of a laptop computer. The MinION™ measures the change in current resulting from DNA strands interacting with a charged protein nanopore. These measurements can then be used to deduce the underlying nucleotide sequence.
Findings: We present a read dataset from whole-genome shotgun sequencing of the model organism Escherichia coli K-12 substr. MG1655 generated on a MinION™ device during the early-access MinION™ Access Program (MAP). Sequencing runs of the MinION™ are presented, one generated using R7 chemistry (released in July 2014) and one using R7.3 (released in September 2014).
Conclusions: Base-called sequence data are provided to demonstrate the nature of data produced by the MinION™ platform and to encourage the development of customised methods for alignment, consensus and variant calling, de novo assembly and scaffolding. FAST5 files containing event data within the HDF5 container format are provided to assist with the development of improved base-calling methods.
Keywords: Genomics; Nanopore sequencing.
Figures
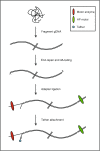
Figure 1
Library preparation schematic for latest version of gDNA sequencing kit (SQK-MAP-003). Figure shows attachment sites for each of the two enzymes and tether.
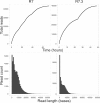
Figure 2
The top row plots demonstrate collector’s curves of sequence reads over time measured in hours for the R7 (left) and R7.3 (right) run. Visible on the R7 run is a change in rate of read acquisition associated with topping-up the flowcell with additional library at hours 12, 24, 36 and 48. For the R7.3 run, the updated MinKNOW software reselects sequencing wells after 24 hours. The bottom row shows the histogram of read counts for reads shorter than 50,000 bp in length for the R7 (left) and R7.3 (right) run.
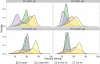
Figure 3
Kernel density plots showing accuracy for R7 and R7.3 chemistries with two different values for the LAST substitution penalty score.
Figure 4
Alignment identity and completeness. Each plot reflects the alignment identity and the proportion of the read aligned for all 2D reads, as well as the underlying template and complement sequences. The top two panels reflect the alignment results for normal and full 2D reads from the R7 flowcell, and the bottom two panels reflect the R7.3 flowcell. Left panels employ a mismatch penalty of 1 and right panels reflect a mismatch penalty of 2. Overall, the lower mismatch penalty increases the identity and fraction of the read that aligned and this effect is greatest for full 2D reads.
Similar articles
- Species-level resolution of 16S rRNA gene amplicons sequenced through the MinION™ portable nanopore sequencer.
Benítez-Páez A, Portune KJ, Sanz Y. Benítez-Páez A, et al. Gigascience. 2016 Jan 28;5:4. doi: 10.1186/s13742-016-0111-z. eCollection 2016. Gigascience. 2016. PMID: 26823973 Free PMC article. - Bacterial and viral identification and differentiation by amplicon sequencing on the MinION nanopore sequencer.
Kilianski A, Haas JL, Corriveau EJ, Liem AT, Willis KL, Kadavy DR, Rosenzweig CN, Minot SS. Kilianski A, et al. Gigascience. 2015 Mar 26;4:12. doi: 10.1186/s13742-015-0051-z. eCollection 2015. Gigascience. 2015. PMID: 25815165 Free PMC article. - Multi-locus and long amplicon sequencing approach to study microbial diversity at species level using the MinION™ portable nanopore sequencer.
Benítez-Páez A, Sanz Y. Benítez-Páez A, et al. Gigascience. 2017 Jul 1;6(7):1-12. doi: 10.1093/gigascience/gix043. Gigascience. 2017. PMID: 28605506 Free PMC article. - Nanopore sequencing: Review of potential applications in functional genomics.
Kono N, Arakawa K. Kono N, et al. Dev Growth Differ. 2019 Jun;61(5):316-326. doi: 10.1111/dgd.12608. Epub 2019 Apr 29. Dev Growth Differ. 2019. PMID: 31037722 Review. - [Nanopore Sequencing and its Application in Biology].
Żmieńko A, Satyr A. Żmieńko A, et al. Postepy Biochem. 2020 Aug 9;66(3):193-204. doi: 10.18388/pb.2020_328. Print 2020 Sep 30. Postepy Biochem. 2020. PMID: 33315321 Review. Polish.
Cited by
- Early insights into the potential of the Oxford Nanopore MinION for the detection of antimicrobial resistance genes.
Judge K, Harris SR, Reuter S, Parkhill J, Peacock SJ. Judge K, et al. J Antimicrob Chemother. 2015 Oct;70(10):2775-8. doi: 10.1093/jac/dkv206. Epub 2015 Jul 28. J Antimicrob Chemother. 2015. PMID: 26221019 Free PMC article. - Improved data analysis for the MinION nanopore sequencer.
Jain M, Fiddes IT, Miga KH, Olsen HE, Paten B, Akeson M. Jain M, et al. Nat Methods. 2015 Apr;12(4):351-6. doi: 10.1038/nmeth.3290. Epub 2015 Feb 16. Nat Methods. 2015. PMID: 25686389 Free PMC article. - Perspectives and Benefits of High-Throughput Long-Read Sequencing in Microbial Ecology.
Tedersoo L, Albertsen M, Anslan S, Callahan B. Tedersoo L, et al. Appl Environ Microbiol. 2021 Aug 11;87(17):e0062621. doi: 10.1128/AEM.00626-21. Epub 2021 Aug 11. Appl Environ Microbiol. 2021. PMID: 34132589 Free PMC article. Review. - MinION takes center stage.
Rusk N. Rusk N. Nat Methods. 2015 Jan;12(1):12-3. doi: 10.1038/nmeth.3244. Nat Methods. 2015. PMID: 25699315 No abstract available. - Estimating error rates for single molecule protein sequencing experiments.
Smith MB, VanderVelden K, Blom T, Stout HD, Mapes JH, Folsom TM, Martin C, Bardo AM, Marcotte EM. Smith MB, et al. PLoS Comput Biol. 2024 Jul 5;20(7):e1012258. doi: 10.1371/journal.pcbi.1012258. eCollection 2024 Jul. PLoS Comput Biol. 2024. PMID: 38968291 Free PMC article.
References
- Baldarelli R, Branton D, Church G, Deamer DW, Kasianowicz J. Characterization of individual polymer molecules based on monomer-interface interactions. Google Patents. US Patent 5,795,782. 1998.
- Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–1153. doi: 10.1038/nbt.1495. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources