UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches - PubMed (original) (raw)
UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches
Baris E Suzek et al. Bioinformatics. 2015.
Abstract
Motivation: UniRef databases provide full-scale clustering of UniProtKB sequences and are utilized for a broad range of applications, particularly similarity-based functional annotation. Non-redundancy and intra-cluster homogeneity in UniRef were recently improved by adding a sequence length overlap threshold. Our hypothesis is that these improvements would enhance the speed and sensitivity of similarity searches and improve the consistency of annotation within clusters.
Results: Intra-cluster molecular function consistency was examined by analysis of Gene Ontology terms. Results show that UniRef clusters bring together proteins of identical molecular function in more than 97% of the clusters, implying that clusters are useful for annotation and can also be used to detect annotation inconsistencies. To examine coverage in similarity results, BLASTP searches against UniRef50 followed by expansion of the hit lists with cluster members demonstrated advantages compared with searches against UniProtKB sequences; the searches are concise (∼7 times shorter hit list before expansion), faster (∼6 times) and more sensitive in detection of remote similarities (>96% recall at e-value <0.0001). Our results support the use of UniRef clusters as a comprehensive and scalable alternative to native sequence databases for similarity searches and reinforces its reliability for use in functional annotation.
© The Author 2014. Published by Oxford University Press.
Figures
Fig. 1.
The categories of UniRef clusters based on intra-cluster functional consistency. Upper left panel shows an example of GO term hierarchy used. Other panels illustrate the UniRef clusters in categories based on their intra-cluster consistency; I (All members have identical GO terms), II-1 (all members share common GO terms and some have additional less or equally specific GO terms, not children of the shared GO terms), II-2 (all members share common GO terms and some have additional more specific GO terms), III (only some members share common GO terms but all member’s GO terms can be traced to a common non-root parent GO term, is a child of one of the shared GO terms) and IV (members do not have any common GO term and the existing ones cannot be traced to a common non-root parent GO term)
Fig. 2.
Example UniProtKB/Swiss-Prot (query) and UniProtKB (target) pairs for distant similarity detection analysis, where Pfam domains common to query and targets span more than 80% of target protein sequences
Fig. 3.
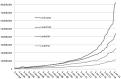
Growth of UniRef databases and UniProt Knowledgebase
Fig. 4.
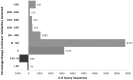
The size distribution of UniRef clusters follows a power law distribution
Fig. 5.
Distribution of UniRef90 clusters specificity for complete set of clusters (top bars) and those containing only model organisms (bottom bars). UniRef50 clusters follow similar distribution
Fig. 6.
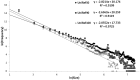
Precision and recall (Equations (2) and (3)) of UniRef50-based BLASTP searches expanded using cluster memberships at different _e_-value thresholds
Fig. 7.
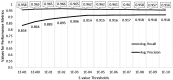
The percentage difference in distant similarities detected by UniRef50- versus UniProtKB-based searches based on the dataset constructed using Pfam domains
Fig. 8.
ROC50 values for UniRef50- versus UniProtKB-based searches based on the dataset constructed using Pfam domains
Similar articles
- UniRef: comprehensive and non-redundant UniProt reference clusters.
Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. Suzek BE, et al. Bioinformatics. 2007 May 15;23(10):1282-8. doi: 10.1093/bioinformatics/btm098. Epub 2007 Mar 22. Bioinformatics. 2007. PMID: 17379688 - Uniclust databases of clustered and deeply annotated protein sequences and alignments.
Mirdita M, von den Driesch L, Galiez C, Martin MJ, Söding J, Steinegger M. Mirdita M, et al. Nucleic Acids Res. 2017 Jan 4;45(D1):D170-D176. doi: 10.1093/nar/gkw1081. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899574 Free PMC article. - Comparison of topological clustering within protein networks using edge metrics that evaluate full sequence, full structure, and active site microenvironment similarity.
Leuthaeuser JB, Knutson ST, Kumar K, Babbitt PC, Fetrow JS. Leuthaeuser JB, et al. Protein Sci. 2015 Sep;24(9):1423-39. doi: 10.1002/pro.2724. Epub 2015 Aug 18. Protein Sci. 2015. PMID: 26073648 Free PMC article. - The Universal Protein Resource (UniProt): an expanding universe of protein information.
Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O'Donovan C, Redaschi N, Suzek B. Wu CH, et al. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D187-91. doi: 10.1093/nar/gkj161. Nucleic Acids Res. 2006. PMID: 16381842 Free PMC article. - Protein function prediction: towards integration of similarity metrics.
Erdin S, Lisewski AM, Lichtarge O. Erdin S, et al. Curr Opin Struct Biol. 2011 Apr;21(2):180-8. doi: 10.1016/j.sbi.2011.02.001. Epub 2011 Feb 24. Curr Opin Struct Biol. 2011. PMID: 21353529 Free PMC article. Review.
Cited by
- Cholesterol Metabolism by Uncultured Human Gut Bacteria Influences Host Cholesterol Level.
Kenny DJ, Plichta DR, Shungin D, Koppel N, Hall AB, Fu B, Vasan RS, Shaw SY, Vlamakis H, Balskus EP, Xavier RJ. Kenny DJ, et al. Cell Host Microbe. 2020 Aug 12;28(2):245-257.e6. doi: 10.1016/j.chom.2020.05.013. Epub 2020 Jun 15. Cell Host Microbe. 2020. PMID: 32544460 Free PMC article. - PredictONCO: a web tool supporting decision-making in precision oncology by extending the bioinformatics predictions with advanced computing and machine learning.
Stourac J, Borko S, Khan RT, Pokorna P, Dobias A, Planas-Iglesias J, Mazurenko S, Pinto G, Szotkowska V, Sterba J, Slaby O, Damborsky J, Bednar D. Stourac J, et al. Brief Bioinform. 2023 Nov 22;25(1):bbad441. doi: 10.1093/bib/bbad441. Brief Bioinform. 2023. PMID: 38066711 Free PMC article. - A comprehensive review and comparison of existing computational methods for protein function prediction.
Lin B, Luo X, Liu Y, Jin X. Lin B, et al. Brief Bioinform. 2024 May 23;25(4):bbae289. doi: 10.1093/bib/bbae289. Brief Bioinform. 2024. PMID: 39003530 Free PMC article. Review. - SPOT: A machine learning model that predicts specific substrates for transport proteins.
Kroll A, Niebuhr N, Butler G, Lercher MJ. Kroll A, et al. PLoS Biol. 2024 Sep 26;22(9):e3002807. doi: 10.1371/journal.pbio.3002807. eCollection 2024 Sep. PLoS Biol. 2024. PMID: 39325691 Free PMC article. - Biochemical characterization of a SusD-like protein involved in β-1,3-glucan utilization by an uncultured cow rumen Bacteroides.
Li X, Lippens G, Parrou J-L, Cioci G, Esque J, Wang Z, Laville E, Potocki-Veronese G, Labourel A. Li X, et al. mSphere. 2024 Aug 28;9(8):e0027824. doi: 10.1128/msphere.00278-24. Epub 2024 Jul 16. mSphere. 2024. PMID: 39012103 Free PMC article.
References
- Altschul S.F., et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. - PubMed
- Cameron M., et al. (2007) Clustered sequence representation for fast homology search. J. Comput. Biol., 14, 594–614. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases