DoGSD: the dog and wolf genome SNP database - PubMed (original) (raw)
. 2015 Jan;43(Database issue):D777-83.
doi: 10.1093/nar/gku1174. Epub 2014 Nov 17.
Wen-Ming Zhao 2, Bi-Xia Tang 3, Yan-Qing Wang 2, Lu Wang 4, Zhang Zhang 5, He-Chuan Yang 6, Yan-Hu Liu 4, Jun-Wei Zhu 2, David M Irwin 7, Guo-Dong Wang 8, Ya-Ping Zhang 9
Affiliations
- PMID: 25404132
- PMCID: PMC4383968
- DOI: 10.1093/nar/gku1174
DoGSD: the dog and wolf genome SNP database
Bing Bai et al. Nucleic Acids Res. 2015 Jan.
Abstract
The rapid advancement of next-generation sequencing technology has generated a deluge of genomic data from domesticated dogs and their wild ancestor, grey wolves, which have simultaneously broadened our understanding of domestication and diseases that are shared by humans and dogs. To address the scarcity of single nucleotide polymorphism (SNP) data provided by authorized databases and to make SNP data more easily/friendly usable and available, we propose DoGSD (http://dogsd.big.ac.cn), the first canidae-specific database which focuses on whole genome SNP data from domesticated dogs and grey wolves. The DoGSD is a web-based, open-access resource comprising ∼ 19 million high-quality whole-genome SNPs. In addition to the dbSNP data set (build 139), DoGSD incorporates a comprehensive collection of SNPs from two newly sequenced samples (1 wolf and 1 dog) and collected SNPs from three latest dog/wolf genetic studies (7 wolves and 68 dogs), which were taken together for analysis with the population genetic statistics, Fst. In addition, DoGSD integrates some closely related information including SNP annotation, summary lists of SNPs located in genes, synonymous and non-synonymous SNPs, sampling location and breed information. All these features make DoGSD a useful resource for in-depth analysis in dog-/wolf-related studies.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Figure 1.
The data sources and pipeline to construct DoGSD.
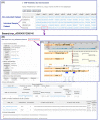
Figure 2.
The screen dumps of the SNP lists of DoGSD and typical browse results. The curly arrow indicates two steps are not directly linked and the straight arrow means two steps are directly linked. (A) The non-redundant and individual sample SNP lists. (B) The text format browse result of an SNP. (C) GBrowse visualization of a SNP.
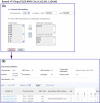
Figure 3.
Example of a comparative search. (A) An example of search entry. (B) The comparative search result.
Similar articles
- Whole genome resequencing of the Iranian native dogs and wolves to unravel variome during dog domestication.
Amiri Ghanatsaman Z, Wang GD, Asadollahpour Nanaei H, Asadi Fozi M, Peng MS, Esmailizadeh A, Zhang YP. Amiri Ghanatsaman Z, et al. BMC Genomics. 2020 Mar 4;21(1):207. doi: 10.1186/s12864-020-6619-8. BMC Genomics. 2020. PMID: 32131720 Free PMC article. - Worldwide patterns of genomic variation and admixture in gray wolves.
Fan Z, Silva P, Gronau I, Wang S, Armero AS, Schweizer RM, Ramirez O, Pollinger J, Galaverni M, Ortega Del-Vecchyo D, Du L, Zhang W, Zhang Z, Xing J, Vilà C, Marques-Bonet T, Godinho R, Yue B, Wayne RK. Fan Z, et al. Genome Res. 2016 Feb;26(2):163-73. doi: 10.1101/gr.197517.115. Epub 2015 Dec 17. Genome Res. 2016. PMID: 26680994 Free PMC article. - The IGF1 small dog haplotype is derived from Middle Eastern grey wolves.
Gray MM, Sutter NB, Ostrander EA, Wayne RK. Gray MM, et al. BMC Biol. 2010 Feb 24;8:16. doi: 10.1186/1741-7007-8-16. BMC Biol. 2010. PMID: 20181231 Free PMC article. - Evolutionary genomics of dog domestication.
Wayne RK, vonHoldt BM. Wayne RK, et al. Mamm Genome. 2012 Feb;23(1-2):3-18. doi: 10.1007/s00335-011-9386-7. Epub 2012 Jan 22. Mamm Genome. 2012. PMID: 22270221 Review. - Deciphering the Origin of Dogs: From Fossils to Genomes.
Freedman AH, Wayne RK. Freedman AH, et al. Annu Rev Anim Biosci. 2017 Feb 8;5:281-307. doi: 10.1146/annurev-animal-022114-110937. Epub 2016 Nov 28. Annu Rev Anim Biosci. 2017. PMID: 27912242 Review.
Cited by
- Comprehensive Analysis of Microsatellite Instability in Canine Cancers: Implications for Comparative Oncology and Personalized Veterinary Medicine.
Mazzone E, Aresu L. Mazzone E, et al. Animals (Basel). 2024 Aug 27;14(17):2484. doi: 10.3390/ani14172484. Animals (Basel). 2024. PMID: 39272269 Free PMC article. - Clinical, pathologic and molecular findings in 2 Rottweiler littermates with appendicular osteosarcoma.
Silver KI, Mannheimer JD, Saba C, Hendricks WPD, Wang G, Day K, Warrier M, Beck JA, Mazcko C, LeBlanc AK. Silver KI, et al. Res Sq [Preprint]. 2024 Apr 11:rs.3.rs-4223759. doi: 10.21203/rs.3.rs-4223759/v1. Res Sq. 2024. PMID: 38659878 Free PMC article. Preprint. - Shared hotspot mutations in oncogenes position dogs as an unparalleled comparative model for precision therapeutics.
Rodrigues L, Watson J, Feng Y, Lewis B, Harvey G, Post G, Megquier K, White ME, Lambert L, Miller A, Lopes C, Zhao S. Rodrigues L, et al. Sci Rep. 2023 Jul 6;13(1):10935. doi: 10.1038/s41598-023-37505-2. Sci Rep. 2023. PMID: 37414794 Free PMC article. - Dog-human translational genomics: state of the art and genomic resources.
Pallotti S, Piras IS, Marchegiani A, Cerquetella M, Napolioni V. Pallotti S, et al. J Appl Genet. 2022 Dec;63(4):703-716. doi: 10.1007/s13353-022-00721-z. Epub 2022 Sep 8. J Appl Genet. 2022. PMID: 36074326 Review. - The genomic landscape of canine diffuse large B-cell lymphoma identifies distinct subtypes with clinical and therapeutic implications.
Giannuzzi D, Marconato L, Fanelli A, Licenziato L, De Maria R, Rinaldi A, Rotta L, Rouquet N, Birolo G, Fariselli P, Mensah AA, Bertoni F, Aresu L. Giannuzzi D, et al. Lab Anim (NY). 2022 Jul;51(7):191-202. doi: 10.1038/s41684-022-00998-x. Epub 2022 Jun 20. Lab Anim (NY). 2022. PMID: 35726023
References
- Savolainen P., Zhang Y.P., Luo J., Lundeberg J., Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science (New York, N.Y.) 2002;298:1610–1613. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous