Genetic basis for clinical response to CTLA-4 blockade in melanoma - PubMed (original) (raw)
. 2014 Dec 4;371(23):2189-2199.
doi: 10.1056/NEJMoa1406498. Epub 2014 Nov 19.
Vladimir Makarov # 1, Taha Merghoub # 1, Jianda Yuan # 1, Jesse M Zaretsky 1, Alexis Desrichard 1, Logan A Walsh 1, Michael A Postow 1, Phillip Wong 1, Teresa S Ho 1, Travis J Hollmann 1, Cameron Bruggeman 1, Kasthuri Kannan 1, Yanyun Li 1, Ceyhan Elipenahli 1, Cailian Liu 1, Christopher T Harbison 1, Lisu Wang 1, Antoni Ribas 1, Jedd D Wolchok # 1, Timothy A Chan # 1
Affiliations
- PMID: 25409260
- PMCID: PMC4315319
- DOI: 10.1056/NEJMoa1406498
Genetic basis for clinical response to CTLA-4 blockade in melanoma
Alexandra Snyder et al. N Engl J Med. 2014.
Erratum in
Abstract
Background: Immune checkpoint inhibitors are effective cancer treatments, but molecular determinants of clinical benefit are unknown. Ipilimumab and tremelimumab are antibodies against cytotoxic T-lymphocyte antigen 4 (CTLA-4). Anti-CTLA-4 treatment prolongs overall survival in patients with melanoma. CTLA-4 blockade activates T cells and enables them to destroy tumor cells.
Methods: We obtained tumor tissue from patients with melanoma who were treated with ipilimumab or tremelimumab. Whole-exome sequencing was performed on tumors and matched blood samples. Somatic mutations and candidate neoantigens generated from these mutations were characterized. Neoantigen peptides were tested for the ability to activate lymphocytes from ipilimumab-treated patients.
Results: Malignant melanoma exomes from 64 patients treated with CTLA-4 blockade were characterized with the use of massively parallel sequencing. A discovery set consisted of 11 patients who derived a long-term clinical benefit and 14 patients who derived a minimal benefit or no benefit. Mutational load was associated with the degree of clinical benefit (P=0.01) but alone was not sufficient to predict benefit. Using genomewide somatic neoepitope analysis and patient-specific HLA typing, we identified candidate tumor neoantigens for each patient. We elucidated a neoantigen landscape that is specifically present in tumors with a strong response to CTLA-4 blockade. We validated this signature in a second set of 39 patients with melanoma who were treated with anti-CTLA-4 antibodies. Predicted neoantigens activated T cells from the patients treated with ipilimumab.
Conclusions: These findings define a genetic basis for benefit from CTLA-4 blockade in melanoma and provide a rationale for examining exomes of patients for whom anti-CTLA-4 agents are being considered. (Funded by the Frederick Adler Fund and others.).
Figures
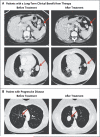
Figure 1. Paired Pretreatment and Post-Treatment Computed Tomographic Scans
In Panel A, the scans on the top were obtained on January 2, 2011, and August 26, 2013, and the scans on the bottom were obtained on September 6, 2011, and January 14, 2013. In Panel B, the scans were obtained on August 13, 2009, and January 9, 2010.
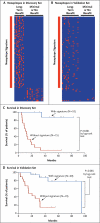
Figure 2. Mutational Landscape of Tumors According to Clinical Benefit from Ipilimumab Treatment
Panel A shows the mutational load (number of nonsynonymous mutations per exome) in the discovery and validation sets, according to status with respect to a clinical benefit from therapy. Panel B depicts the Kaplan–Meier curves for overall survival in the discovery set for patients with more than 100 nonsynonymous coding mutations per exome and patients with 100 or fewer mutations.
Figure 3. Association of a Neoepitope Signature with a Clinical Benefit from CTLA-4 Blockade
Candidate neoepitopes were identified by means of mutational analysis, as described in the Methods section in the Supplementary Appendix. Panel A shows a heat map of candidate tetrapeptide neoantigens that were present in patients with a long-term clinical benefit but absent in patients with a minimal benefit or no benefit in the discovery set (comprising 25 patients). Each row represents a neoepitope; each column represents a patient. The vertical red line indicates the tetra-peptide signature associated with a response to blockade of cytotoxic T-lymphocyte antigen 4 (CTLA-4). The exact tetrapeptides, chromosomal loci, and non-mutant and mutant nonamers in which they occur are listed in Table S6 in the Supplementary Appendix. Panel B shows the same information for the validation set (comprising 39 patients). Panel C shows the Kaplan–Meier curves for overall survival in the discovery set for patients with the signature and those without the signature. Panel D shows the same data for the validation set.
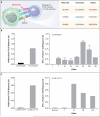
Figure 4. Role of Neoantigens in Activation of T Cells from Patients Treated with CTLA-4 Blockade
Panel A shows an example of a tetrapeptide substring of human cytomegalovirus. In each case, the nonamer containing the mutation is predicted to bind and be presented by a patient-specific HLA. Panel B shows the dual positive (interferon-γ [IFN-γ] and tumor necrosis factor α [TNF-α]) CD8+ T-cell response to T_E_SPFEQHI and nonmutant peptide T_K_SPFEQHI and the increase in IFN-γ+ T cells over time. Data from Patient CR9306 are shown. T bars indicate the standard deviation. Panel C shows the dual positive (IFN-γ and TNF-α) CD8+ T-cell response to GLER_E_GFTF and nonmutant peptide GLER_G_GFTF and illustrates the increase in peptide-specific T cells 24 weeks after the initiation of treatment with ipilimumab relative to baseline. Data from Patient CR0095 are shown. MHC denotes major histocompatibility complex, and TCR T-cell receptor.
Comment in
- Somatic mutations and immunotherapy outcome with CTLA-4 blockade in melanoma.
Boussiotis VA. Boussiotis VA. N Engl J Med. 2014 Dec 4;371(23):2230-2. doi: 10.1056/NEJMe1413061. Epub 2014 Nov 19. N Engl J Med. 2014. PMID: 25409261 Free PMC article. No abstract available. - Skin cancer. Benefiting from CTLA-4 blockade--a genetic rationale.
Errico A. Errico A. Nat Rev Clin Oncol. 2015 Jan;12(1):4. doi: 10.1038/nrclinonc.2014.217. Epub 2014 Dec 9. Nat Rev Clin Oncol. 2015. PMID: 25488393 No abstract available. - Genetic basis for clinical response to CTLA-4 blockade.
Snyder A, Wolchok JD, Chan TA. Snyder A, et al. N Engl J Med. 2015 Feb 19;372(8):783. doi: 10.1056/NEJMc1415938. N Engl J Med. 2015. PMID: 25693024 No abstract available. - Genetic basis for clinical response to CTLA-4 blockade.
Melero I, Lasarte JJ. Melero I, et al. N Engl J Med. 2015 Feb 19;372(8):783. doi: 10.1056/NEJMc1415938. N Engl J Med. 2015. PMID: 25693025 No abstract available. - Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma.
Chan TA, Wolchok JD, Snyder A. Chan TA, et al. N Engl J Med. 2015 Nov 12;373(20):1984. doi: 10.1056/NEJMc1508163. N Engl J Med. 2015. PMID: 26559592 No abstract available.
Similar articles
- Genomic correlates of response to CTLA-4 blockade in metastatic melanoma.
Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA. Van Allen EM, et al. Science. 2015 Oct 9;350(6257):207-211. doi: 10.1126/science.aad0095. Epub 2015 Sep 10. Science. 2015. PMID: 26359337 Free PMC article. - Prognostic factors and outcomes in metastatic uveal melanoma treated with programmed cell death-1 or combined PD-1/cytotoxic T-lymphocyte antigen-4 inhibition.
Heppt MV, Heinzerling L, Kähler KC, Forschner A, Kirchberger MC, Loquai C, Meissner M, Meier F, Terheyden P, Schell B, Herbst R, Göppner D, Kiecker F, Rafei-Shamsabadi D, Haferkamp S, Huber MA, Utikal J, Ziemer M, Bumeder I, Pfeiffer C, Schäd SG, Schmid-Tannwald C, Tietze JK, Eigentler TK, Berking C. Heppt MV, et al. Eur J Cancer. 2017 Sep;82:56-65. doi: 10.1016/j.ejca.2017.05.038. Epub 2017 Jun 22. Eur J Cancer. 2017. PMID: 28648699 - Targeting immune checkpoints in unresectable metastatic cutaneous melanoma: a systematic review and meta-analysis of anti-CTLA-4 and anti-PD-1 agents trials.
Yun S, Vincelette ND, Green MR, Wahner Hendrickson AE, Abraham I. Yun S, et al. Cancer Med. 2016 Jul;5(7):1481-91. doi: 10.1002/cam4.732. Epub 2016 May 11. Cancer Med. 2016. PMID: 27167347 Free PMC article. Review. - Ipilimumab: a novel immunostimulatory monoclonal antibody for the treatment of cancer.
Graziani G, Tentori L, Navarra P. Graziani G, et al. Pharmacol Res. 2012 Jan;65(1):9-22. doi: 10.1016/j.phrs.2011.09.002. Epub 2011 Sep 10. Pharmacol Res. 2012. PMID: 21930211 Review. - Immune checkpoint blockade.
Naidoo J, Page DB, Wolchok JD. Naidoo J, et al. Hematol Oncol Clin North Am. 2014 Jun;28(3):585-600. doi: 10.1016/j.hoc.2014.02.002. Hematol Oncol Clin North Am. 2014. PMID: 24880949 Review.
Cited by
- Eligibility and Endpoints for Clinical Trials in Trimodality Therapy for Bladder Cancer.
Singh P, Ballas L, Sonpavde GP, Chen RC, Bangs R, Bauman BC, Nagar H, Delacroix SE, Lerner SP, Efstathiou JA. Singh P, et al. Bladder Cancer. 2024 Oct 23;10(3):199-213. doi: 10.3233/BLC-240036. eCollection 2024. Bladder Cancer. 2024. PMID: 39493817 Free PMC article. - A comprehensive investigation of associations between cell death pathways and molecular and clinical features in pan-cancer.
He Y, Wang X. He Y, et al. Clin Transl Oncol. 2024 Nov 2. doi: 10.1007/s12094-024-03769-x. Online ahead of print. Clin Transl Oncol. 2024. PMID: 39487950 - Gut microbiome metabolites, molecular mimicry, and species-level variation drive long-term efficacy and adverse event outcomes in lung cancer survivors.
Liu X, Lu B, Tang H, Jia X, Zhou Q, Zeng Y, Gao X, Chen M, Xu Y, Wang M, Tan B, Li J. Liu X, et al. EBioMedicine. 2024 Oct 28;109:105427. doi: 10.1016/j.ebiom.2024.105427. Online ahead of print. EBioMedicine. 2024. PMID: 39471749 Free PMC article. - Peripheral blood lymphocyte subpopulations as predictive biomarkers for first-line programmed death 1 inhibitors efficacy in esophageal squamous cell carcinoma: A retrospective study.
Sun J, Gan W, Yao J, Han Z, Fang Z, Xiong W, Li D, Wu J, Cao L, Zhu L. Sun J, et al. Medicine (Baltimore). 2024 Oct 4;103(40):e39967. doi: 10.1097/MD.0000000000039967. Medicine (Baltimore). 2024. PMID: 39465723 Free PMC article. - Construction of a tumor mutational burden-derived LncRNA prognostic computational framework associated with therapy sensitivity in skin cutaneous melanoma.
Li G, Wu T, Li H, Wei C, Sun Y, Gao P, Huang X, Liu Z, Li J, Wang Y, Li G, Fan L. Li G, et al. J Transl Med. 2024 Oct 24;22(1):966. doi: 10.1186/s12967-024-05732-4. J Transl Med. 2024. PMID: 39449143 Free PMC article.
References
- Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. [Erratum, N Engl J Med 2010;363:1290.]
- Gajewski TF, Louahed J, Brichard VG. Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. Cancer J. 2010;16:399–403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials