CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer - PubMed (original) (raw)
. 2014 Nov 20;159(5):1126-1139.
doi: 10.1016/j.cell.2014.10.024. Epub 2014 Nov 6.
Eugenio Marco 2, Camilla L Christensen 3, Nicholas Kwiatkowski 4, Tinghu Zhang 5, Clark M Hatheway 6, Brian J Abraham 4, Bandana Sharma 6, Caleb Yeung 1, Abigail Altabef 3, Antonio Perez-Atayde 7, Kwok-Kin Wong 3, Guo-Cheng Yuan 2, Nathanael S Gray 5, Richard A Young 4, Rani E George 8
Affiliations
- PMID: 25416950
- PMCID: PMC4243043
- DOI: 10.1016/j.cell.2014.10.024
CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer
Edmond Chipumuro et al. Cell. 2014.
Abstract
The MYC oncoproteins are thought to stimulate tumor cell growth and proliferation through amplification of gene transcription, a mechanism that has thwarted most efforts to inhibit MYC function as potential cancer therapy. Using a covalent inhibitor of cyclin-dependent kinase 7 (CDK7) to disrupt the transcription of amplified MYCN in neuroblastoma cells, we demonstrate downregulation of the oncoprotein with consequent massive suppression of MYCN-driven global transcriptional amplification. This response translated to significant tumor regression in a mouse model of high-risk neuroblastoma, without the introduction of systemic toxicity. The striking treatment selectivity of MYCN-overexpressing cells correlated with preferential downregulation of super-enhancer-associated genes, including MYCN and other known oncogenic drivers in neuroblastoma. These results indicate that CDK7 inhibition, by selectively targeting the mechanisms that promote global transcriptional amplification in tumor cells, may be useful therapy for cancers that are driven by MYC family oncoproteins.
Figures
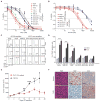
Figure 1. THZ1 Exhibits High Potency and Selectivity Against _MYCN_-amplified Tumor Cells
(A) Dose-response curves of _MYCN_-amplified and nonamplified human NB and murine fibroblast cells after treatment with increasing concentrations of THZ1 for 72 hr. Percent cell viability relative to that of DMSO-treated cells is shown here and in (B). Data represent mean ± SD of 3 replicates here and in (B). (B) Dose-response curves of NB cells treated as in (A) with the reversible CDK7 inhibitor THZ1R. (C) Cell-cycle analysis of _MYCN_-amplified vs. nonamplified NB cells exposed to THZ1 (100 nM × 24 and 48 hr) by flow cytometry with propidium iodide (PI) staining. Results are representative of 3 replicates. The scale and axes are indicated in the lower left corner. (D) Apoptosis analysis in _MYCN_-amplified and nonamplified NB cells treated with THZ1 as in (C) by flow cytometry with Annexin V staining. Data represent mean ± SD of 3 replicates. ***p<0.0001, **p<0.001 (Student’s t-test). (E) Tumor volumes of _MYCN_-amplified human NB xenografts in NU/NU (Crl:NU-Foxn1nu) mice treated with THZ1 (10 mg/kg IV twice daily) (n=14) or vehicle (n=9) for 28 days. Mean ± SD values are presented. ***p<0.001; **p<0.01; *p<0.05 (multiple t-test, Holm-Sidak method). (F) Immunohistochemical (IHC) analysis of morphology (hematoxylin & eosin, H&E), proliferation (Ki67) and apoptosis (cleaved caspase 3, CC3) in tumors harvested from animals treated with vehicle or THZ1 as in (E) for 12 days. Scale bar, 25 μM. See also Figure S1, Table S1.
Figure 2. THZ1 Inhibits General Transcription and Cell Cycle Regulation in _MYCN_-amplified Tumor Models
(A) Immunoblot analysis of RNA Pol II CTD phosphorylation in_MYCN_-amplified and nonamplified NB cells treated with DMSO or THZ1 at the indicated concentrations for the indicated times. (B) Immunoblot analysis of RNA Pol II CTD phosphorylation in human NB xenograft tumor cells obtained from mice treated with vehicle or THZ1 (10 mg/kg IV twice daily) for 12 days. (C) Immunoblot analysis of MCL1 in _MYCN_-amplified (Kelly, NGP) vs. nonamplified (SK-N-FI, SH-SY5Y) NB cells following treatment with THZ1 at the indicated concentrations and durations. (D) Immunoblot analysis of proteins involved in cell cycle progression in_MYCN_-amplified (Kelly, IMR-32) and nonamplified (SH-SY5Y) NB cells following treatment with THZ1 100 nM for 3 and 6 hr. See also Figure S2
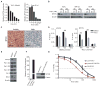
Figure 3. THZ1 Causes Massive Downregulation of Actively Transcribed Genes in _MYCN_-overexpressing NB Cells
(A) Heat map of gene expression values in _MYCN_-amplified and nonamplified cells treated with THZ1 (100 nM for 6 hr) vs. DMSO. Rows show z-scores calculated for each cell type. (B) Quartile box plots of log2 fold changes in gene expression in_MYCN_-amplified and nonamplified cells treated with DMSO or THZ1 at the same dose and duration as in (A). Box plot whiskers extend to 1.5 times the interquartile range (n=18,665 expressed genes, p<10−15 for Kelly vs. SH-SY5Y and IMR-32 vs. SH-SY5Y, two-sided Mann-Whitney U test). (C) Venn diagram depicting the overlap between sets of differentially expressed transcripts (THZ1 vs. DMSO) in _MYCN_-amplified (Kelly, IMR-32) and nonamplified (SH-SY5Y) cells treated with THZ1 as in (A). Red represents upregulated and blue, downregulated transcripts. (D) Heat map of gene expression values of transcriptional and cell cycle CDKs in_MYCN_-amplified and nonamplified cells treated with THZ1 as in (A) vs. DMSO. Rows show z-scores calculated for each cell type. (E) Correlation between log2 fold changes in gene expression following THZ1 (100 nM) vs. DMSO treatment and actinomycin D (1 μM) vs. DMSO treatment for 6 hr. in _MYCN_-amplified NB cells. R2 (coefficient of determination) calculated using a simple linear regression model. See also Figure S3.
Figure 4. Cytotoxic Effects of THZ1 are Mediated in Part Through Inhibition of MYCN Expression
(A) Quantitative RT-PCR (qRT-PCR) analysis of MYCN RNA expression in _MYCN_-amplified Kelly cells treated with THZ1-50 nM for 0–6 hr (left) or 0–250 nM for 6 hr (right). Data normalized to β-actin are presented as mean ± SD of three biological replicates. (B) Immunoblot analysis of MYCN protein expression in_MYCN-_amplified NB cells treated with the indicated doses of THZ1 for 6 or 24 hr. (C) IHC analysis of MYCN protein expression in _MYCN_-amplified human NB xenograft models treated with either DMSO or 10 mg/kg IV twice daily of THZ1 for 12 days. Scale bars, 25 μM. (D) ChIP-qPCR analysis of MYCN binding at the promoters of candidate target genes in _MYCN_-amplified cells following treatment with THZ1, 100 nM for 3 hr. Mean ± SD values for three replicate experiments are shown. **p<0.01 (Student’s t-test). (E) Immunoblot analysis of the indicated proteins in_MYCN_-amplified NB cells expressing either an shRNA control (shGFP) or an shRNA directed against MYCN (shMYCN). Two different hairpins against MYCN [shMYCN(1) and shMYCN(3)] were used with similar results. (F) qRT-PCR analysis of MYCN expression in SH-SY5Y_MYCN_-nonamplified cells engineered to express either a control vector (pLenti 6.3) or MYCN (pLenti 6.3 MYCN (left panel). Immunoblot analysis of MYCN protein expression in these cells (right panel). (G) Cell viability analysis of untransfected (SH-SY5Y), control vector-expressing (pLenti 6.3) or MYCN-expressing (pLenti 6.3 MYCN)_MYCN_-nonamplified NB cells treated for 72 hr with increasing doses of THZ1. Results are means ± SD of 3 replicates. See also Figure S4.
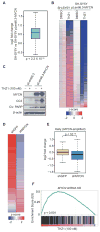
Figure 5. THZ1 Inhibits _MYCN_-driven Transcriptional Amplification
(A) Box plot depicting the log2 fold changes in actively transcribed genes in_MYCN_-nonamplified SH-SY5Y cells transduced with_MYCN_ (pLenti 6.3-MYCN) compared with a control vector (pLenti 6.3). Box plot whiskers extend to 1.5 times the interquartile range (n=18,665 expressed genes; p<2.2 × 10−16, two-sided Mann-Whitney U test). (B) Heat map of all gene expression values in _MYCN_-nonamplified SH-SY5Y cells vs. SH-SY5Y cells overexpressing MYCN (pLenti 6.3-MYCN) following either THZ1 (100 nM for 6 hr) or DMSO treatment. Rows show z-scores calculated for each vector. (C) Immunoblot analysis of MYCN, CC3 and cleaved PARP in SH-SY5Y_MYCN_-nonamplified cells engineered to express either a control vector (pLenti 6.3) or MYCN (pLenti 6.3-MYCN) following treatment with THZ1. (D) Heat map of differentially expressed genes in _MYCN_-amplified NB cells expressing a MYCN shRNA vs. control shRNA. Rows show z-scores calculated for each vector. (E) Box plots of log2 fold changes in gene expression in_MYCN_-amplified NB cells expressing a _MYCN_shRNA vs. a control shRNA. Box plot whiskers extend to 1.5 times the interquartile range (n=18,665 expressed genes; p<10−15 for shMYCN vs. shGFP, two-sided Mann-Whitney U test). (F) GSEA plot depicting the correlation between the top 500 downregulated genes following THZ1 treatment and the rank-ordered genes that are differentially expressed after MYCN knockdown in_MYCN_-amplified NB cells. See also Figure S5.
Figure 6. NB Cells Possess Unique Super-enhancer Landscapes
(A) H3K27ac signal across enhancer regions for all enhancers in_MYCN_-amplified and nonamplified cells. SEs were defined as enhancers surpassing the threshold signal of 8802 in both cell types. In Kelly and SH-SY5Y cells, 6.2% (746/12,000) and 5.4% (1136/20,887) of the enhancers comprised 52% and 37%, respectively of all H3K27ac-bound enhancer signal and were classed as SEs. (B) ChIP-seq profiles for H3K27ac, H3K4me1 and RNA Pol II binding at the_MYCN_ SE gene locus in Kelly cells. The x axis shows genomic position and the y axis the signal of histone mark or Pol II binding in units of reads per million per base pair (rpm/bp). The gene model is depicted below and scale bars above the binding profiles. (C) ChIP-seq profiles for H3K27ac binding at representative SE-associated gene loci in _MYCN_-amplified and nonamplified cells. The x and y axes are as described in (B). (D) Gene Ontology (GO) categories of SE-associated genes in_MYCN_-amplified (Kelly) and nonamplified (SH-SY5Y) cells identified using GREAT analysis (McLean et al., 2010). (E) H3K27ac loading across enhancers in _MYCN_-amplified and nonamplified primary NB tumors. SEs were defined as having a threshold signal of ~5600 in the tumors. In NB#1 and NB#2, 8.5% (164/1,920) and 3.0% (109/3,561) and in NB#3, 6.4% (522/8,040) of the enhancers were classified as SEs. See also Figure S6.
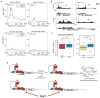
Figure 7. Sensitivity of _MYCN_-amplified Cells to THZ1 Correlates with _MYCN_-associated Super-enhancers
(A) Metagene representation of global Pol II ChIP-seq occupancy at SE- and RE-associated genes in cells with (Kelly) or without (SH-SY5Y)MYCN amplification treated with DMSO (blue) or THZ1 (red) (100 nM × 3 hr). TSS, transcription start sites. (B) Gene tracks of RNA Pol II binding density at representative SE-associated gene loci after DMSO or THZ1 treatment as in (A). (C) Left, quartile box plots of log2 fold changes in the top 300 genes associated with SEs and regular enhancers (RE) in _MYCN_-amplified cells treated with THZ1 (100 nM × 6 hr) vs. DMSO. Box plot whiskers extend to 1.5 times the interquartile range (n=230 SE; n=231 RE; p<10−2, two-sided Mann-Whitney U test). Right, quartile box plots of log2 fold changes in gene transcripts associated with the top-ranked SEs unique to _MYCN_-amplified Kelly cells treated with THZ1 as in (A) vs. DMSO, compared with the expression changes of the same genes in similarly treated SH-SY5Y nonamplified cells. Box plot whiskers extend to 1.5 times the interquartile range (n=673; p<10−16, two-sided Mann-Whitney U test). (D) Proposed mechanism for the action of THZ1 in _MYCN_-amplified NB. Oncogenic MYCN is regulated by super-enhancers leading to its high-level expression (left). Overexpressed MYCN invades the promoter and enhancer regions of all active genes, including itself, to induce global transcriptional upregulation (right). THZ1 targets the expression of both MYCN and _MYCN_-driven transcriptional amplification.
Similar articles
- Combination therapy with the CDK7 inhibitor and the tyrosine kinase inhibitor exerts synergistic anticancer effects against MYCN-amplified neuroblastoma.
Tee AE, Ciampa OC, Wong M, Fletcher JI, Kamili A, Chen J, Ho N, Sun Y, Carter DR, Cheung BB, Marshall GM, Liu PY, Liu T. Tee AE, et al. Int J Cancer. 2020 Oct 1;147(7):1928-1938. doi: 10.1002/ijc.32936. Epub 2020 Mar 16. Int J Cancer. 2020. PMID: 32086952 - CDK7 inhibition suppresses aberrant hedgehog pathway and overcomes resistance to smoothened antagonists.
Liu F, Jiang W, Sui Y, Meng W, Hou L, Li T, Li M, Zhang L, Mo J, Wang J, Zhao Y, Zhang L, Ma J, Tang Y. Liu F, et al. Proc Natl Acad Sci U S A. 2019 Jun 25;116(26):12986-12995. doi: 10.1073/pnas.1815780116. Epub 2019 Jun 10. Proc Natl Acad Sci U S A. 2019. PMID: 31182587 Free PMC article. - CDK/CK1 inhibitors roscovitine and CR8 downregulate amplified MYCN in neuroblastoma cells.
Delehouzé C, Godl K, Loaëc N, Bruyère C, Desban N, Oumata N, Galons H, Roumeliotis TI, Giannopoulou EG, Grenet J, Twitchell D, Lahti J, Mouchet N, Galibert MD, Garbis SD, Meijer L. Delehouzé C, et al. Oncogene. 2014 Dec 11;33(50):5675-87. doi: 10.1038/onc.2013.513. Epub 2013 Dec 9. Oncogene. 2014. PMID: 24317512 Free PMC article. - The MYCN oncoprotein as a drug development target.
Lu X, Pearson A, Lunec J. Lu X, et al. Cancer Lett. 2003 Jul 18;197(1-2):125-30. doi: 10.1016/s0304-3835(03)00096-x. Cancer Lett. 2003. PMID: 12880971 Review. - Cyclin-dependent kinase 7 inhibitors in cancer therapy.
Wang M, Wang T, Zhang X, Wu X, Jiang S. Wang M, et al. Future Med Chem. 2020 May;12(9):813-833. doi: 10.4155/fmc-2019-0334. Epub 2020 Mar 25. Future Med Chem. 2020. PMID: 32208930 Review.
Cited by
- Transcriptional CDK Inhibitors CYC065 and THZ1 Induce Apoptosis in Glioma Stem Cells Derived from Recurrent GBM.
Juric V, Düssmann H, Lamfers MLM, Prehn JHM, Rehm M, Murphy BM. Juric V, et al. Cells. 2021 May 12;10(5):1182. doi: 10.3390/cells10051182. Cells. 2021. PMID: 34066147 Free PMC article. - The molecular understanding of super-enhancer dysregulation in cancer.
Yoshino S, Suzuki HI. Yoshino S, et al. Nagoya J Med Sci. 2022 May;84(2):216-229. doi: 10.18999/nagjms.84.2.216. Nagoya J Med Sci. 2022. PMID: 35967935 Free PMC article. Review. - Molecular mechanisms of resistance to kinase inhibitors and redifferentiation in thyroid cancers.
Hofmann MC, Kunnimalaiyaan M, Wang JR, Busaidy NL, Sherman SI, Lai SY, Zafereo M, Cabanillas ME. Hofmann MC, et al. Endocr Relat Cancer. 2022 Sep 14;29(11):R173-R190. doi: 10.1530/ERC-22-0129. Print 2022 Nov 1. Endocr Relat Cancer. 2022. PMID: 35975971 Free PMC article. Review. - Targeting MYCN and ALK in resistant and relapsing neuroblastoma.
Tucker ER, Poon E, Chesler L. Tucker ER, et al. Cancer Drug Resist. 2019 Sep 19;2(3):803-812. doi: 10.20517/cdr.2019.009. eCollection 2019. Cancer Drug Resist. 2019. PMID: 35582571 Free PMC article. Review. - Characterization of super-enhancer-associated functional lncRNAs acting as ceRNAs in ESCC.
Wang QY, Peng L, Chen Y, Liao LD, Chen JX, Li M, Li YY, Qian FC, Zhang YX, Wang F, Li CQ, Lin DC, Xu LY, Li EM. Wang QY, et al. Mol Oncol. 2020 Sep;14(9):2203-2230. doi: 10.1002/1878-0261.12726. Epub 2020 Jun 20. Mol Oncol. 2020. PMID: 32460441 Free PMC article.
References
- Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16:313–317. - PubMed
- Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. - PubMed
- Cohn SL, Salwen H, Quasney MW, Ikegaki N, Cowan JM, Herst CV, Kennett RH, Rosen ST, DiGiuseppe JA, Brodeur GM. Prolonged N-myc protein half-life in a neuroblastoma cell line lacking N-myc amplification. Oncogene. 1990;5:1821–1827. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA148688S1/CA/NCI NIH HHS/United States
- R21 HG006778/HG/NHGRI NIH HHS/United States
- R21HG006778/HG/NHGRI NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- R01 CA179483/CA/NCI NIH HHS/United States
- R01CA148688/CA/NCI NIH HHS/United States
- R01 CA148688/CA/NCI NIH HHS/United States
- R01 HG002668/HG/NHGRI NIH HHS/United States
- HG002668/HG/NHGRI NIH HHS/United States
- R01CA179483-01/CA/NCI NIH HHS/United States
- CA109901/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials