Autoregulatory mechanism for dynactin control of processive and diffusive dynein transport - PubMed (original) (raw)
. 2014 Dec;16(12):1192-201.
doi: 10.1038/ncb3063. Epub 2014 Nov 24.
Affiliations
- PMID: 25419851
- PMCID: PMC4250405
- DOI: 10.1038/ncb3063
Autoregulatory mechanism for dynactin control of processive and diffusive dynein transport
Suvranta K Tripathy et al. Nat Cell Biol. 2014 Dec.
Abstract
Dynactin is the longest known cytoplasmic dynein regulator, with roles in dynein recruitment to subcellular cargo and in stimulating processive dynein movement. The latter function was thought to involve the N-terminal microtubule-binding region of the major dynactin polypeptide p150(Glued), although recent results disputed this. To understand how dynactin regulates dynein we generated recombinant fragments of the N-terminal half of p150(Glued). We find that the dynein-binding coiled-coil α-helical domain CC1B is sufficient to stimulate dynein processivity, which it accomplishes by increasing average dynein step size and forward-step frequency, while decreasing lateral stepping and microtubule detachment. In contrast, the immediate upstream coiled-coil domain, CC1A, activates a surprising diffusive dynein state. CC1A interacts physically with CC1B and interferes with its effect on dynein processivity. We also identify a role for the N-terminal portion of p150(Glued) in coordinating these activities. Our results reveal an unexpected form of long-range allosteric control of dynein motor function by internal p150(Glued) sequences, and evidence for p150(Glued) autoregulation.
Figures
Figure 1. Characterization of dynactinp150_Glued_ fragments
(A) Diagram of the dynactin complex, an ~35 nm long filament of the actin-like protein Arp1 and associated factors. The p150_Glued_ subunit is seen as a projecting arm at left with globular N-terminal microtubule binding CAP-Gly (green) and basic (orange) domains near the end. (B) Domain map of p150_Glued_ and its subfragments used in this study, which are C-terminally flag (*) and His6 (+) tagged. CC1B and CC1A contain slightly different boundaries from those used in our previous study. (C) Coomassie stained gel of the purified p150_Glued_ fragments used in this study. (3 independent experiments) (D) Calf brain cytoplasmic dynein was tested for co immunoprecipitation with the Flag-tagged p150_Glued_ fragments using anti-flag antibody. Bands were visualized by Western blotting with antibodies to dynein heavy chain and the flag tag. All fragments except CC1A bound dynein. (3 independent experiments) (E) Microtubule (MT) co sedimentation of p150_Glued_ fragments. Fragments (0.1 uM, unless otherwise noted) were centrifuged in the absence or presence of taxol-stabilized MTs. Only p150 1-555, which alone contains the CAP-Gly and complete basic regions of p150_Glued_, showed substantial co-sedimentation with MTs. (2–3 independent experiments) CC: coiled-coil α-helix, Sup: supernatant, Ab: antibody.
Figure 2. Effects of dynactin fragments on dynein single molecule processivity
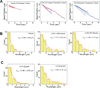
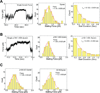
(A) Sample traces (4 each) showing processive motion of beads with dynein alone (left) dynein plus p135-CC1 (middle) and dynein plus CC1B (right). (B) Bead run length distributions for dynein alone (left), dynein with P150 (middle), or dynein with P135 (right). (C) Run length histograms for dynein with CC1 (left) and CC1B (right). In each case, data were well described by a single decaying exponential (chi-squared test) with mean travel for dynein plus the p150_Glued_ 1-555, p135-CC1, and CC1B fragments approximately double that for dynein alone (see also Table 1). All processivity measurements were done at a bead binding fraction of ~30%.
Figure 3. Effects of dynactin fragments on dynein single molecule diffusivity
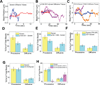
Sample traces (3–4) showing diffusive motion of beads with dynein alone (A), dynein with p135-CC1 (B) and dynein with high amounts of CC1A (C). (D–H) Quantitation of the effects of different dynactin fragments on the overall amount of diffusive vs processive binding events. Because the percentage of dynein-only diffusion could change between experiments, each graph reports the results of the control (dynein-alone) performed at the same time as the fragment experiment. The sum of the diffusive and processive components reflects the total bead binding fraction (e.g. in D, the total bead binding fraction for dynein alone was 30%). P135 (D) and P150 (E) decreased the gross number of processive events, and increased the number of diffusive events. CC1 (F) decreased the gross number of processive events, but did not increase diffusive events, and CC1B (G) did not alter the number of either class of events. CC1A (H) had no effect at low concentrations, but induced diffusion and suppressed processive motion at high concentrations. These data reflect numerous experiments. Error bars (Fig 3 D–H) were obtained using equation p*(1−p)/N, where p refers to bead binding fraction and N refers to the total number of bead tested. For dynein alone, dynein-p135 CC1, dynein-p150 1-555, dynein-CC1B, dynein-CC1 (70% BF), dynein-CC1A (150:1), and for dynein-CC1A (7000:1) the respective number of beads checked was N = 165, 195, 180, 253, 161, 70, and 70. For high dynein (70% BF), N = 85. Each experiment was reproduced in the lab at least 3 times. The single dynein alone control experiment was repeated before each measurement of dynein with a dynactin fragment.
Figure 4. Effects of dynactin fragments on dynein single molecule force production
(A) reflects dynein alone, and (B) reflects dynein with P150. Shown are example force traces (A, B, Left), distributions of stalling forces (100 force events) (A, B, middle) and distributions of durations of force producing events (40 stalling events) (A,B, Right). C) shows distributions of stalling forces for dynein with P135 (left) and CC1B (right).
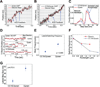
Figure 5. Effects of processivity-enhancing fragment CC1B on dynein stepping behavior
Sample dynein bead traces with detected steps (in red) for single dynein alone (A) and with CC1B (B). Arrows indicate points of force feedback (see Methods). (C) Distribution of step-sizes for single dyneins with (blue) or without (red) CC1B (428 stepping events). CC1B reduces back-stepping (green arrow) and increases large forward steps (blue arrow). (D) Sample traces showing lateral bead position on microtubule vs. time for stationary dynein (without ATP - top); and dynein alone (middle) or with CC1B (bottom) moving on a microtubule in the presence of saturating ATP. Step detection identifies changes in lateral bead position (red lines); δ indicates size of detected lateral step in nm. (E) Lateral dynein bead switching frequency (95 switching events) on the microtubule surface, and (F) as function of run length (94 run lengths); p value from t-test. Longer CC1B-dynein runs correlate with decreased lateral switching frequency. (G) Magnitude of lateral step size (428 detected steps). CC1B decreases the frequency and magnitude of lateral dynein steps. Error bars are SEM.
Figure 6. Interaction between coiled-coil p150_Glued_ fragments CC1A and CC1B
(A) Pull-down of bacterially-expressed CC1B with GST-CC1A or GST alone shows specific binding of CC1B to CC1A using Coomassie Blue staining. (3 independent experiments) (B) Size exclusion FPLC of CC1A and CC1B using Coomassie Blue Staining. (3 independent experiments) When in combination the two fragments elute in a common, higher molecular size peak. (C, D) Single-molecule bead assays reveal that CC1A, at a level which has no effect on its own on dynein (150:1 CC1A:dynein), strongly suppresses the effect of CC1B on dynein (see Figure legend 3 for number of beads checked). The number of processive dynein events and their average run-length are reduced relative to values for dynein alone, suggesting that, as for CC1, the combined fragments actively inhibit dynein function. Error is SEM. E) Schematic representation of dynein regulation by dynactin. (Left) Dimer of p150_Glued_ N-terminus (aa 1-555) binds via its CC1B coiled-coil domain with the dynein intermediate chains situated within the dynein tail. CC1B alone is sufficient to increase the length of processive dynein movement along microtubules. CC1A is shown interacting physically with CC1B. As the two domains are covalently linked in the intact p150_Glued_ and p135 polypeptides, we propose that CC1 must bend as shown. In this conformation, the N-terminal globular portion of p150_Glued_ and p135 might be able to interact with the C-terminal portion of CC1B. Thus, we envision a series of intramolecular interactions within p150_Glued_ or p135, ultimately modulating dynein behavior through its tail domain. We propose that the ultimate target of regulation are the dynein motor domains, the behavior of which seem more clearly coordinated in the presence of the dynactin fragments, as evidenced by more efficient stepping along microtubules and longer runs.
Similar articles
- Dynactin p150 promotes processive motility of DDB complexes by minimizing diffusional behavior of dynein.
Feng Q, Gicking AM, Hancock WO. Feng Q, et al. Mol Biol Cell. 2020 Apr 1;31(8):782-792. doi: 10.1091/mbc.E19-09-0495. Epub 2020 Feb 5. Mol Biol Cell. 2020. PMID: 32023147 Free PMC article. - Microtubule binding by dynactin is required for microtubule organization but not cargo transport.
Kim H, Ling SC, Rogers GC, Kural C, Selvin PR, Rogers SL, Gelfand VI. Kim H, et al. J Cell Biol. 2007 Feb 26;176(5):641-51. doi: 10.1083/jcb.200608128. J Cell Biol. 2007. PMID: 17325206 Free PMC article. - Multivalency, autoinhibition, and protein disorder in the regulation of interactions of dynein intermediate chain with dynactin and the nuclear distribution protein.
Jara KA, Loening NM, Reardon PN, Yu Z, Woonnimani P, Brooks C, Vesely CH, Barbar EJ. Jara KA, et al. Elife. 2022 Nov 23;11:e80217. doi: 10.7554/eLife.80217. Elife. 2022. PMID: 36416224 Free PMC article. - [Dynein and dynactin as organizers of the system of cell microtubules].
Burakov AV, Nadezhdina ES. Burakov AV, et al. Ontogenez. 2006 Sep-Oct;37(5):323-39. Ontogenez. 2006. PMID: 17066975 Review. Russian. - Regulation of dynein-dynactin-driven vesicular transport.
Liu JJ. Liu JJ. Traffic. 2017 Jun;18(6):336-347. doi: 10.1111/tra.12475. Epub 2017 Mar 28. Traffic. 2017. PMID: 28248450 Review.
Cited by
- Regulation of in vivo dynein force production by CDK5 and 14-3-3ε and KIAA0528.
Chapman DE, Reddy BJN, Huy B, Bovyn MJ, Cruz SJS, Al-Shammari ZM, Han H, Wang W, Smith DS, Gross SP. Chapman DE, et al. Nat Commun. 2019 Jan 16;10(1):228. doi: 10.1038/s41467-018-08110-z. Nat Commun. 2019. PMID: 30651536 Free PMC article. - Control of cytoplasmic dynein force production and processivity by its C-terminal domain.
Nicholas MP, Höök P, Brenner S, Wynne CL, Vallee RB, Gennerich A. Nicholas MP, et al. Nat Commun. 2015 Feb 11;6:6206. doi: 10.1038/ncomms7206. Nat Commun. 2015. PMID: 25670086 Free PMC article. - Step Sizes and Rate Constants of Single-headed Cytoplasmic Dynein Measured with Optical Tweezers.
Kinoshita Y, Kambara T, Nishikawa K, Kaya M, Higuchi H. Kinoshita Y, et al. Sci Rep. 2018 Nov 5;8(1):16333. doi: 10.1038/s41598-018-34549-7. Sci Rep. 2018. PMID: 30397249 Free PMC article. - Dynactin p150 promotes processive motility of DDB complexes by minimizing diffusional behavior of dynein.
Feng Q, Gicking AM, Hancock WO. Feng Q, et al. Mol Biol Cell. 2020 Apr 1;31(8):782-792. doi: 10.1091/mbc.E19-09-0495. Epub 2020 Feb 5. Mol Biol Cell. 2020. PMID: 32023147 Free PMC article. - Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Lau CK, O'Reilly FJ, Santhanam B, Lacey SE, Rappsilber J, Carter AP. Lau CK, et al. EMBO J. 2021 Apr 15;40(8):e106164. doi: 10.15252/embj.2020106164. Epub 2021 Mar 18. EMBO J. 2021. PMID: 33734450 Free PMC article.
References
- Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nat Cell Biol. 2012;14:224–230. - PubMed
- Schroer TA. Dynactin. Annu Rev Cell Dev Bi. 2004;20:759–779. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM070676/GM/NIGMS NIH HHS/United States
- GM102347/GM/NIGMS NIH HHS/United States
- T32 GM008798/GM/NIGMS NIH HHS/United States
- R01 GM070676/GM/NIGMS NIH HHS/United States
- R01 GM102347/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases