Identification of prolyl hydroxylation modifications in mammalian cell proteins - PubMed (original) (raw)
Identification of prolyl hydroxylation modifications in mammalian cell proteins
Patrick R Arsenault et al. Proteomics. 2015 Apr.
Abstract
Prolyl hydroxylation is a PTM that plays an important role in the formation of collagen fibrils and in the oxygen-dependent regulation of hypoxia inducible factor-α (HIF-α). While this modification has been well characterized in the context of these proteins, it remains unclear to what extent it occurs in the remaining mammalian proteome. We explored this question using MS to analyze cellular extracts subjected to various fractionation strategies. In one strategy, we employed the von Hippel Lindau tumor suppressor protein, which recognizes prolyl hydroxylated HIF-α, as a scaffold for generating hydroxyproline capture reagents. We report novel sites of prolyl hydroxylation within five proteins: FK506-binding protein 10, myosin heavy chain 10, hexokinase 2, pyruvate kinase, and C-1 Tetrahydrofolate synthase. Furthermore, we show that identification of prolyl hydroxylation presents a significant technical challenge owing to widespread isobaric methionine oxidation, and that manual inspection of spectra of modified peptides in this context is critical for validation.
Keywords: Cell biology; MS; PTM; Prolyl hydroxylase domain protein; Prolyl hydroxylation.
© 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflicts of Interest: The authors declare no conflict of interest.
Figures
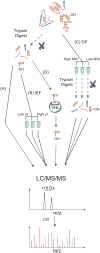
Figure 1
Outline of sample preparation and enrichment strategies. (A) Direct analysis of unfractionated tryptic digests. (B) Following tryptic digests but prior to LC injection, peptides are fractionated by isoelectric focusing (IEF). (C) Fractionation of intact proteins by gel filtration chromatography, followed by tryptic digest prior to LC injection (GF). (D) VHL capture probe-based enrichment of hydroxylated peptides.
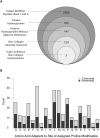
Figure 2
(A) Outline of results of computational analyses. Diagram shows results of Tide-based hydroxyproline search. Largest circle indicates the set of unique peptides that are called with at least one oxidation event (i.e. Proline, Methionine, etc.) as either the most likely, or when applicable, the second most likely candidate for a given spectra (i.e. first and second ranked peptide by XCorr score). Successive filtering steps are indicated. (B) Patterns of enrichment for amino acids immediately adjacent to primary site of assigned prolyl hydroxylation. Values presented are derived from the set of 185 unique peptides identified by Tide analyses as being likely prolyl hydroxylated.
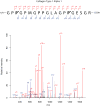
Figure 3
Representative fragmentation spectra (MS2) for Collagen type I, α1 chain. The b and y ions are indicated only when ions appeared above a 2-fold signal to noise ratio. Ions appearing in +2 charge states are indicated with “++”; all others are singly charged species.
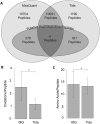
Figure 4
(A) Comparison of overall peptide and putative hydroxyproline peptide identification rates between Tide- and MaxQuant-based analyses platforms at the 1% FDR level. All values refer to numbers of unique peptides. “Putative Hydroxyproline Peptides” refers to the total set of peptides identified as being prolyl hydroxylated by the given analysis package prior to subtraction of likely confounding methionine oxidation and/or manual inspection of fragment spectra. Comparison of (B) average number of oxidations per unique modified peptide and (C) average length of both modified and unmodified peptides, as identified by MaxQuant and Tide analyses respectively. Bars indicate +/- 1 SD, and * indicates p < 0.05 by Student's t-Test.
Similar articles
- Proline hydroxylation at different sites in hypoxia-inducible factor 1α modulates its interactions with the von Hippel-Lindau tumor suppressor protein.
Qian H , Zou Y , Tang Y , Gong Y , Qian Z , Wei G , Zhang Q . Qian H , et al. Phys Chem Chem Phys. 2018 Jul 11;20(27):18756-18765. doi: 10.1039/c8cp01964a. Phys Chem Chem Phys. 2018. PMID: 29961792 - C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation.
Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. Epstein AC, et al. Cell. 2001 Oct 5;107(1):43-54. doi: 10.1016/s0092-8674(01)00507-4. Cell. 2001. PMID: 11595184 - Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation.
Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Jaakkola P, et al. Science. 2001 Apr 20;292(5516):468-72. doi: 10.1126/science.1059796. Epub 2001 Apr 5. Science. 2001. PMID: 11292861 - Protein Hydroxylation by Hypoxia-Inducible Factor (HIF) Hydroxylases: Unique or Ubiquitous?
Strowitzki MJ, Cummins EP, Taylor CT. Strowitzki MJ, et al. Cells. 2019 Apr 26;8(5):384. doi: 10.3390/cells8050384. Cells. 2019. PMID: 31035491 Free PMC article. Review. - Regulation of HIF by the von Hippel-Lindau tumour suppressor: implications for cellular oxygen sensing.
Mole DR, Maxwell PH, Pugh CW, Ratcliffe PJ. Mole DR, et al. IUBMB Life. 2001 Jul;52(1-2):43-7. doi: 10.1080/15216540252774757. IUBMB Life. 2001. PMID: 11795592 Review.
Cited by
- Von Hippel-Lindau regulates interleukin-32β stability in ovarian cancer cells.
Yong HJ, Park JS, Lee Jeong A, Han S, Lee S, Ka HI, Sumiyasuren B, Joo HJ, So SJ, Park JY, Yoon DY, Lim JS, Lee MS, Lee HG, Yang Y. Yong HJ, et al. Oncotarget. 2017 Jul 17;8(41):69833-69846. doi: 10.18632/oncotarget.19311. eCollection 2017 Sep 19. Oncotarget. 2017. PMID: 29050245 Free PMC article. - HypDB: A functionally annotated web-based database of the proline hydroxylation proteome.
Gong Y, Behera G, Erber L, Luo A, Chen Y. Gong Y, et al. PLoS Biol. 2022 Aug 26;20(8):e3001757. doi: 10.1371/journal.pbio.3001757. eCollection 2022 Aug. PLoS Biol. 2022. PMID: 36026437 Free PMC article. - Species Identification of Bovine, Ovine and Porcine Type 1 Collagen; Comparing Peptide Mass Fingerprinting and LC-Based Proteomics Methods.
Buckley M. Buckley M. Int J Mol Sci. 2016 Mar 24;17(4):445. doi: 10.3390/ijms17040445. Int J Mol Sci. 2016. PMID: 27023524 Free PMC article. - Hypoxia-Modified Cancer Cell Metabolism.
Al Tameemi W, Dale TP, Al-Jumaily RMK, Forsyth NR. Al Tameemi W, et al. Front Cell Dev Biol. 2019 Jan 29;7:4. doi: 10.3389/fcell.2019.00004. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 30761299 Free PMC article. Review. - Proteomic analysis reveals diverse proline hydroxylation-mediated oxygen-sensing cellular pathways in cancer cells.
Zhou T, Erber L, Liu B, Gao Y, Ruan HB, Chen Y. Zhou T, et al. Oncotarget. 2016 Nov 29;7(48):79154-79169. doi: 10.18632/oncotarget.12632. Oncotarget. 2016. PMID: 27764789 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources