Granules harboring translationally active mRNAs provide a platform for P-body formation following stress - PubMed (original) (raw)
Granules harboring translationally active mRNAs provide a platform for P-body formation following stress
Jennifer Lui et al. Cell Rep. 2014.
Abstract
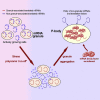
The localization of mRNA to defined cytoplasmic sites in eukaryotic cells not only allows localized protein production but also determines the fate of mRNAs. For instance, translationally repressed mRNAs localize to P-bodies and stress granules where their decay and storage, respectively, are directed. Here, we find that several mRNAs are localized to granules in unstressed, actively growing cells. These granules play a key role in the stress-dependent formation of P-bodies. Specific glycolytic mRNAs are colocalized in multiple granules per cell, which aggregate during P-body formation. Such aggregation is still observed under conditions or in mutants where P-bodies do not form. In unstressed cells, the mRNA granules appear associated with active translation; this might enable a coregulation of protein expression from the same pathways or complexes. Parallels can be drawn between this coregulation and the advantage of operons in prokaryotic systems.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
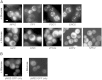
Figure 1
Specific mRNAs Localize into Granules in Unstressed Cells (A) Epifluorescent microscopic z stack images of exponential cells expressing endogenous 3′ UTR MS2L-tagged mRNAs visualized via coexpressed MS2-GFP3. (B) Images of controls expressing pMS2-GFP3 (ymk1741) in either SCD (+glucose) or SC (−glucose) media. Cells were grown to exponential phase in SCD (+glucose) media prior to imaging. Scale bars, 2 μm. See also Figure S1 and Table S1.
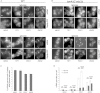
Figure 2
Localization of mRNAs in Granules Does Not Rely on P-Body Formation but Rather Recruits P-Body Components and Coalesce following Glucose Depletion Epifluorescent images of cells grown to exponential phase and then incubated in either SCD (+glucose) or SC media (−glucose) for 10 min. (A) Images of wild-type (WT) cells expressing the P-body marker Dcp2p-CFP as well as MS2L-tagged mRNAs visualized with pMS2-GFP3. (B) Images from mutant strains deficient in P-body formation (edc3Δ lsm4ΔC) also expressing Dcp2p-CFP, MS2-tagged mRNA, and pMS2-GFP3. Scale bar, 2 μm. (C and D) Bar charts showing (C) the percentage of mRNA granules colocalized with P-bodies (Dcp2p) following glucose starvation and (D) the mean number of mRNA granules per cell in wild-type (WT) and edc3Δ lsm4ΔC mutants under glucose replete and starvation conditions. z stack merged images were used to count granules across 50 cells. Errors bars are ±SE. See also Figure S2.
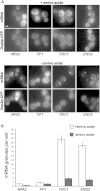
Figure 3
Other Stresses Cause mRNA Granule Aggregation without Inducing P-Bodies Epifluorescent images of cells grown to exponential phase and then incubated in either SCD (+amino acids) or SC-AAs (−amino acids) for 10 min. (A) Images of cells expressing Dcp2p-CFP and MS2L-tagged mRNA/pMS2-GFP3. Scale bar, 2 μm. (B) Quantification of the mean number of mRNA granules per cell under unstressed and amino acid starvation conditions. Merged z stacks were used to count the granules from at least 50 cells. Errors bars are ±SE.
Figure 4
The Recruitment of mRNA Decay Factors to Form P-Bodies Occurs on Preexisting mRNA Granules Epifluorescent images of cells expressing Dcp2p-CFP and MS2L-tagged ENO2 pMS2-GFP3 growing in a microfluidic chamber where the media has been switched for glucose free media and images of cells are collected every minute. The triangle and diamonds highlight mRNA granules, which serve as sites of P-body formation. See also Figure S3.
Figure 5
Cycloheximide Treatment Inhibits the Coalescence of mRNA Granules (A) Schematic representing the culture treatment regimen. Gray and white bars represent 10 min growth in media with or without glucose respectively, and arrowheads denote the point of cycloheximide addition. (B) Epifluorescent images of cells expressing Dcp2-CFP and MS2L-mRNA/pMS2-GFP3. Cells were grown to exponential phase in media containing glucose and then either (1) treated with cycloheximide for 10 min or (2) treated with cycloheximide for 10 min and then incubated in media lacking glucose for 10 min. (C) Bar chart depicting quantification of the average mRNA granules per cell for the treatments described above. Merged z stacks were used to count the number of granules in 50 cells. Errors bars are ±SE. See also Figure S4.
Figure 6
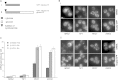
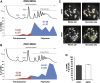
Most PDC1 and ENO2 mRNAs Are Associated with Polysomes and Localized to mRNA Granules (A and B) Polysome fractionation and qRT-PCR analysis on RNA prepared from individual fractions across polysome gradients. Polysomes were analyzed as described in Experimental Procedures. Traces depicting the changes in A254 across the gradient from the yMK1577 (ENO2-MS2L) and yMK1586 (PDC1-MS2L) strains grown in YPD are shown. The 40S (small ribosomal subunit), 60S (large ribosomal subunit), 80S (monosome), and polysome peaks are labeled. Below the percentage of each mRNA present in the fractions collected from the polysome gradient is plotted. Blue represents RNA in polysomal regions, whereas red is from the subpolysomal regions of the gradient. The total percentage in polysomal and subpolysomal regions across three repeat experiments is also depicted. (C and D) (C) Representative images depicting the strategy for quantitating the percentage of PDC1 and ENO2 mRNAs in granules. Fluorescence was measured for the whole cell and granules only (as defined within the yellow lines), and the percentage of each mRNA present in the granules was calculated and plotted in (D). Scale bar, 1 μm. Errors bars are ±SE. See also Figure S4.
Figure 7
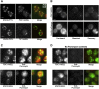
PDC1 and ENO2 mRNAs Are Translated in the Same mRNA Granules (A) Epifluorescent microscopic z stack images of exponential cells expressing endogenous PP7L-tagged ENO2 (visualized via PP7-GFP2, left images) and either MS2L-tagged PDC1 or MS2L-tagged TIF1 (visualized via coexpressed MS2-mCherry3, middle images). Merged images are shown (right) with the percentage GFP granules overlapping with mCherry granules quantified across 50 cells for ENO2 v PDC1. No colocalization was observed for ENO2 v TIF1. Error bar is ±SE. (B) Figure shows a FRAP experiment on the Eno2p-mOrange bearing strain yMK1993 where recovery after photobleaching is followed relative to the localization of the PDC1-MS2L mRNA (visualized using MS2-GFP3). Prebleach, bleached, and recovery images are shown for PDC1 mRNA (top row) and mOrange-tagged Eno2p protein (bottom row). (C) Immunofluoresence using an antipuromycin antibody on cells treated with puromycin/cycloheximide to trap puromycin at the site of protein synthesis (center panels). The MS2-GFP3 mRNA signal for PDC1 and ENO2 is maintained during the procedure (left panels). Merged images show the overlap of the puromycin signal with the mRNA granules (right panels). (D) As in (C), except puromycin was omitted from the procedure. Scale bars, 2 μm throughout. See also Figures S5 and S6.
Similar articles
- Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth.
Pizzinga M, Bates C, Lui J, Forte G, Morales-Polanco F, Linney E, Knotkova B, Wilson B, Solari CA, Berchowitz LE, Portela P, Ashe MP. Pizzinga M, et al. J Cell Biol. 2019 May 6;218(5):1564-1581. doi: 10.1083/jcb.201704019. Epub 2019 Mar 15. J Cell Biol. 2019. PMID: 30877141 Free PMC article. - Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae.
Yoon JH, Choi EJ, Parker R. Yoon JH, et al. J Cell Biol. 2010 May 31;189(5):813-27. doi: 10.1083/jcb.200912019. J Cell Biol. 2010. PMID: 20513766 Free PMC article. - Relationship of GW/P-bodies with stress granules.
Stoecklin G, Kedersha N. Stoecklin G, et al. Adv Exp Med Biol. 2013;768:197-211. doi: 10.1007/978-1-4614-5107-5_12. Adv Exp Med Biol. 2013. PMID: 23224972 Free PMC article. Review. - Yeast mRNA localization: protein asymmetry, organelle localization and response to stress.
Pizzinga M, Ashe MP. Pizzinga M, et al. Biochem Soc Trans. 2014 Aug;42(4):1256-60. doi: 10.1042/BST20140086. Biochem Soc Trans. 2014. PMID: 25110034 Review.
Cited by
- A working model for condensate RNA-binding proteins as matchmakers for protein complex assembly.
Chen X, Mayr C. Chen X, et al. RNA. 2022 Jan;28(1):76-87. doi: 10.1261/rna.078995.121. Epub 2021 Oct 27. RNA. 2022. PMID: 34706978 Free PMC article. Review. - Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth.
Pizzinga M, Bates C, Lui J, Forte G, Morales-Polanco F, Linney E, Knotkova B, Wilson B, Solari CA, Berchowitz LE, Portela P, Ashe MP. Pizzinga M, et al. J Cell Biol. 2019 May 6;218(5):1564-1581. doi: 10.1083/jcb.201704019. Epub 2019 Mar 15. J Cell Biol. 2019. PMID: 30877141 Free PMC article. - Is there a localized role for translational quality control?
Meydan S, Guydosh NR. Meydan S, et al. RNA. 2023 Nov;29(11):1623-1643. doi: 10.1261/rna.079683.123. Epub 2023 Aug 15. RNA. 2023. PMID: 37582617 Free PMC article. - The Role of RNA in Biological Phase Separations.
Fay MM, Anderson PJ. Fay MM, et al. J Mol Biol. 2018 Nov 2;430(23):4685-4701. doi: 10.1016/j.jmb.2018.05.003. Epub 2018 May 10. J Mol Biol. 2018. PMID: 29753780 Free PMC article. Review. - RNA Binding Proteins in Intestinal Epithelial Biology and Colorectal Cancer.
Chatterji P, Rustgi AK. Chatterji P, et al. Trends Mol Med. 2018 May;24(5):490-506. doi: 10.1016/j.molmed.2018.03.008. Epub 2018 Apr 5. Trends Mol Med. 2018. PMID: 29627433 Free PMC article. Review.
References
- Anand M., Chakraburtty K., Marton M.J., Hinnebusch A.G., Kinzy T.G. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J. Biol. Chem. 2003;278:6985–6991. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/K005979/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 099732/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- BB/K005979 /1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G012571/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G0125711/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 088141/Z/09/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases