Cross-tissue and tissue-specific eQTLs: partitioning the heritability of a complex trait - PubMed (original) (raw)
Cross-tissue and tissue-specific eQTLs: partitioning the heritability of a complex trait
Jason M Torres et al. Am J Hum Genet. 2014.
Abstract
Top signals from genome-wide association studies (GWASs) of type 2 diabetes (T2D) are enriched with expression quantitative trait loci (eQTLs) identified in skeletal muscle and adipose tissue. We therefore hypothesized that such eQTLs might account for a disproportionate share of the heritability estimated from all SNPs interrogated through GWASs. To test this hypothesis, we applied linear mixed models to the Wellcome Trust Case Control Consortium (WTCCC) T2D data set and to data sets representing Mexican Americans from Starr County, TX, and Mexicans from Mexico City. We estimated the proportion of phenotypic variance attributable to the additive effect of all variants interrogated in these GWASs, as well as a much smaller set of variants identified as eQTLs in human adipose tissue, skeletal muscle, and lymphoblastoid cell lines. The narrow-sense heritability explained by all interrogated SNPs in each of these data sets was substantially greater than the heritability accounted for by genome-wide-significant SNPs (∼10%); GWAS SNPs explained over 50% of phenotypic variance in the WTCCC, Starr County, and Mexico City data sets. The estimate of heritability attributable to cross-tissue eQTLs was greater in the WTCCC data set and among lean Hispanics, whereas adipose eQTLs significantly explained heritability among Hispanics with a body mass index ≥ 30. These results support an important role for regulatory variants in the genetic component of T2D susceptibility, particularly for eQTLs that elicit effects across insulin-responsive peripheral tissues.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
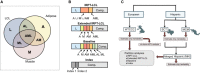
Figure 1
Study Overview (A) Delineation of the eQTL subsets evaluated in the decomposition of T2D heritability. (B) Diagram showing the constitution of each of the major partitions evaluated in this study. (C) Schematic of the heritability analyses performed on the WTCCC, SCT, and MCM data sets.
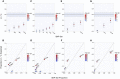
Figure 2
Estimates of Narrow-Sense T2D Heritability Explained by GWAS-Interrogated SNPs (A) The REML estimates of phenotypic variance explained by the additive effect of SNPs interrogated in GWASs (V A / V P) on T2D are shown for the WTCCC, SCT, and MCM data sets. (B–D) Heritability estimates for SNP subsets composed of T2D-associated variants from the NHGRI GWAS catalog and HapMap2 SNPs within 1 kb, 10 kb, 100 kb, 500 kb, and 1Mb are shown for the WTCCC (B), SCT (C), and MCM (D) data sets. Total chip heritability and SE for each GWAS are given by the solid and dashed black lines, respectively. The color corresponds to the significance of each heritability estimate determined by the test statistic from the likelihood-ratio test (LRT).
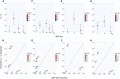
Figure 3
Heritability of T2D Explained by Metabolic-Tissue eQTLs in the WTCCC GWAS Data Set The narrow-sense heritability estimates (V A / V P) attributable to nonoverlapping SNP subsets (top panels). The proportion of chip heritability explained by each subset is plotted with SNP-set proportion (bottom panels). Color is designated by the −log10 of the LRT p value, and estimates are shown with SE. (A and B) IRPT-LCL analysis. (C and D) Expanded IRPT-LCL analysis. (E and F) Index-subset analysis with muscle-specific (M) and cross-tissue (CT) eQTLs as index sets. (G and H) _cis_-trans analysis of cross-tissue eQTLs.
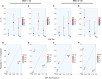
Figure 4
Heritability of T2D Explained by Metabolic-Tissue eQTLs in the Merged Hispanic Data Set The narrow-sense heritability estimates (V A / V P) attributable to nonoverlapping SNP subsets (top panels). The proportion of chip heritability explained by each subset is plotted with SNP-set proportion (bottom panels). Color is designated by the −log10 of the LRT p value, and estimates are shown with SE. (A and B) IRPT-LCL analysis. (C and D) Expanded IRPT-LCL analysis. (E and F) Index-subset analysis with muscle-specific (M) and cross-tissue (CT) eQTLs as index sets. (G and H) Index-subset analysis with adipose-specific (A) and cross-tissue (CT) eQTLs as index sets.
Figure 5
Partition of Metabolic-Tissue Heritability among Hispanics in Low- and High-BMI Subgroups The narrow-sense heritability estimates (V A / V P) attributable to nonoverlapping SNP subsets are shown with SE and color corresponding to LRT (top panels). The proportion of chip heritability explained by each subset is plotted with SNP-set proportion and is color coded by the −log10 of the LRT p value (bottom panels). Results from an index-subset analysis with muscle-specific (M) and cross-tissue (CT) eQTLs as the index sets are shown for Hispanic subjects with a BMI < 30 (A and B) and subjects with a BMI ≥ 30 (E and F). Results from an index-subset analysis with adipose-specific (A) and cross-tissue (CT) eQTLs as the index sets are shown for Hispanic subjects with a BMI < 30 (C and D) and subjects with a BMI ≥ 30 (F and G).
Similar articles
- Genome-wide association and meta-analysis in populations from Starr County, Texas, and Mexico City identify type 2 diabetes susceptibility loci and enrichment for expression quantitative trait loci in top signals.
Below JE, Gamazon ER, Morrison JV, Konkashbaev A, Pluzhnikov A, McKeigue PM, Parra EJ, Elbein SC, Hallman DM, Nicolae DL, Bell GI, Cruz M, Cox NJ, Hanis CL. Below JE, et al. Diabetologia. 2011 Aug;54(8):2047-55. doi: 10.1007/s00125-011-2188-3. Epub 2011 Jun 7. Diabetologia. 2011. PMID: 21647700 Free PMC article. - Mapping adipose and muscle tissue expression quantitative trait loci in African Americans to identify genes for type 2 diabetes and obesity.
Sajuthi SP, Sharma NK, Chou JW, Palmer ND, McWilliams DR, Beal J, Comeau ME, Ma L, Calles-Escandon J, Demons J, Rogers S, Cherry K, Menon L, Kouba E, Davis D, Burris M, Byerly SJ, Ng MC, Maruthur NM, Patel SR, Bielak LF, Lange LA, Guo X, Sale MM, Chan KH, Monda KL, Chen GK, Taylor K, Palmer C, Edwards TL, North KE, Haiman CA, Bowden DW, Freedman BI, Langefeld CD, Das SK. Sajuthi SP, et al. Hum Genet. 2016 Aug;135(8):869-80. doi: 10.1007/s00439-016-1680-8. Epub 2016 May 19. Hum Genet. 2016. PMID: 27193597 Free PMC article. - A systematic heritability analysis of the human whole blood transcriptome.
Huan T, Liu C, Joehanes R, Zhang X, Chen BH, Johnson AD, Yao C, Courchesne P, O'Donnell CJ, Munson PJ, Levy D. Huan T, et al. Hum Genet. 2015 Mar;134(3):343-58. doi: 10.1007/s00439-014-1524-3. Epub 2015 Jan 14. Hum Genet. 2015. PMID: 25585846 Free PMC article. - Genetic Component of Type 2 Diabetes in a Mexican Population.
Sánchez-Pozos K, Menjívar M. Sánchez-Pozos K, et al. Arch Med Res. 2016 Oct;47(7):496-505. doi: 10.1016/j.arcmed.2016.12.007. Arch Med Res. 2016. PMID: 28262190 Review. - Shared genetic etiology underlying Alzheimer's disease and type 2 diabetes.
Hao K, Di Narzo AF, Ho L, Luo W, Li S, Chen R, Li T, Dubner L, Pasinetti GM. Hao K, et al. Mol Aspects Med. 2015 Jun-Oct;43-44:66-76. doi: 10.1016/j.mam.2015.06.006. Epub 2015 Jun 23. Mol Aspects Med. 2015. PMID: 26116273 Free PMC article. Review.
Cited by
- Leveraging lung tissue transcriptome to uncover candidate causal genes in COPD genetic associations.
Lamontagne M, Bérubé JC, Obeidat M, Cho MH, Hobbs BD, Sakornsakolpat P, de Jong K, Boezen HM; International COPD Genetics Consortium; Nickle D, Hao K, Timens W, van den Berge M, Joubert P, Laviolette M, Sin DD, Paré PD, Bossé Y. Lamontagne M, et al. Hum Mol Genet. 2018 May 15;27(10):1819-1829. doi: 10.1093/hmg/ddy091. Hum Mol Genet. 2018. PMID: 29547942 Free PMC article. - A Perspective of the Cross-Tissue Interplay of Genetics, Epigenetics, and Transcriptomics, and Their Relation to Brain Based Phenotypes in Schizophrenia.
Liu J, Chen J, Perrone-Bizzozero N, Calhoun VD. Liu J, et al. Front Genet. 2018 Aug 23;9:343. doi: 10.3389/fgene.2018.00343. eCollection 2018. Front Genet. 2018. PMID: 30190726 Free PMC article. Review. - Quantifying genetic effects on disease mediated by assayed gene expression levels.
Yao DW, O'Connor LJ, Price AL, Gusev A. Yao DW, et al. Nat Genet. 2020 Jun;52(6):626-633. doi: 10.1038/s41588-020-0625-2. Epub 2020 May 18. Nat Genet. 2020. PMID: 32424349 Free PMC article. - Accuracy of Gene Expression Prediction From Genotype Data With PrediXcan Varies Across and Within Continental Populations.
Mikhaylova AV, Thornton TA. Mikhaylova AV, et al. Front Genet. 2019 Apr 3;10:261. doi: 10.3389/fgene.2019.00261. eCollection 2019. Front Genet. 2019. PMID: 31001318 Free PMC article. - Survey of the Heritability and Sparse Architecture of Gene Expression Traits across Human Tissues.
Wheeler HE, Shah KP, Brenner J, Garcia T, Aquino-Michaels K; GTEx Consortium; Cox NJ, Nicolae DL, Im HK. Wheeler HE, et al. PLoS Genet. 2016 Nov 11;12(11):e1006423. doi: 10.1371/journal.pgen.1006423. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27835642 Free PMC article.
References
- Poulsen P., Kyvik K.O., Vaag A., Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—a population-based twin study. Diabetologia. 1999;42:139–145. - PubMed
- Kaprio J., Tuomilehto J., Koskenvuo M., Romanov K., Reunanen A., Eriksson J., Stengård J., Kesäniemi Y.A. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. 1992;35:1060–1067. - PubMed
- Newman B., Selby J.V., King M.C., Slemenda C., Fabsitz R., Friedman G.D. Concordance for type 2 (non-insulin-dependent) diabetes mellitus in male twins. Diabetologia. 1987;30:763–768. - PubMed
- Lyssenko V., Almgren P., Anevski D., Perfekt R., Lahti K., Nissén M., Isomaa B., Forsen B., Homström N., Saloranta C., Botnia study group Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes. 2005;54:166–174. - PubMed
- Almgren P., Lehtovirta M., Isomaa B., Sarelin L., Taskinen M.R., Lyssenko V., Tuomi T., Groop L., Botnia Study Group Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia. 2011;54:2811–2819. - PubMed
Publication types
MeSH terms
Grants and funding
- F31 DK101202/DK/NIDDK NIH HHS/United States
- TL1 TR000432/TR/NCATS NIH HHS/United States
- CAPMC/ CIHR/Canada
- R01 HL102830/HL/NHLBI NIH HHS/United States
- HHSN268200782096C/HL/NHLBI NIH HHS/United States
- R01 MH101820/MH/NIMH NIH HHS/United States
- R01 MH090937/MH/NIMH NIH HHS/United States
- P30 DK020595/DK/NIDDK NIH HHS/United States
- U01 DK085501/DK/NIDDK NIH HHS/United States
- UL1 TR000430/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical