Genome-wide RNAi screen reveals a role for multipass membrane proteins in endosome-to-golgi retrieval - PubMed (original) (raw)
Genome-wide RNAi screen reveals a role for multipass membrane proteins in endosome-to-golgi retrieval
Sophia Y Breusegem et al. Cell Rep. 2014.
Abstract
Endosome-to-Golgi retrieval is an essential membrane trafficking pathway required for many important physiological processes and linked to neurodegenerative disease and infection by bacterial and viral pathogens. The prototypical cargo protein for this pathway is the cation-independent mannose 6-phosphate receptor (CIMPR), which delivers lysosomal hydrolases to endosomes. Efficient retrieval of CIMPR to the Golgi requires the retromer complex, but other aspects of the endosome-to-Golgi retrieval pathway are poorly understood. Employing an image-based antibody-uptake assay, we conducted a genome-wide RNAi loss-of-function screen for novel regulators of this trafficking pathway and report ∼90 genes that are required for endosome-to-Golgi retrieval of a CD8-CIMPR reporter protein. Among these regulators of endosome-to-Golgi retrieval are a number of multipass membrane-spanning proteins, a class of proteins often overlooked with respect to a role in membrane trafficking. We further demonstrate a role for three multipass membrane proteins, SFT2D2, ZDHHC5, and GRINA, in endosome-to-Golgi retrieval.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
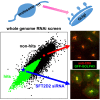
Graphical abstract
Figure 1
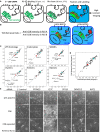
Pilot Screen for Known Regulators of Endosome-to-Golgi Retrieval Using Anti-CD8 Uptake Assay (A) Schematic of the anti-CD8 antibody retrieval assay as adapted for high-throughput screening. HeLa cells stably expressing a CD8-CIMPR reporter and the Golgi protein GFP-GOLPH3 were used. Anti-CD8 antibody (Ab) was bound at room temperature for 15 min and chased at 37°C for 30 min. Labeling and imaging details are in Supplemental Experimental Procedures. (B) Definition of the TGN retrieval ratio used in our studies and depiction of subcellular antibody localizations that give rise to high or low TGN retrieval ratios. (C) Scatterplots of the TGN retrieval ratios measured in replicate screens of 60 VPS gene homologs, 60 endocytosis genes, 43 SNARE protein genes, 41 dynein genes, 49 kinesin genes, and 43 myosin genes. Negative control measurements are in blue; SNX1 siRNA positive control measurements are in red. A linear fit to the data points is also shown. Marked data points indicate known endosome-to-Golgi retrieval pathway genes or genes that were selected for further validation. (D) Replicate TGN retrieval ratios for the validation screen of the pilot study, in which 80 individual ON TARGETplus siRNA sequences (four per gene) were assayed. Controls and linear fit are as in (C). (E) Representative images of the pilot validation screen for the negative control (no siRNA) and for five sets of siRNA-treated cells with reduced TGN retrieval ratio. Different phenotypes that give rise to reduced TGN retrieval ratios are discussed in the text. Scale bar, 100 μm (top two rows) and 38 μm (bottom row).
Figure 2
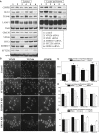
A Whole-Genome siRNA Screen to Identify Regulators of Endosome-to-Golgi Retrieval (A) Plate layout for genome-wide loss-of-function screen. (B) Whole-genome screen hit selection. (C) Scatterplot of the normalized TGN retrieval ratios for the two replicates of the whole-genome screen. Each data point represents a single siRNA pool. Only valid measurements (18,465 siRNA pools) are included. Negative (cyan square) and positive (SNX1 siRNA, red data points) controls are shown. Green data points indicate siRNA pools with TGN retrieval ratio SSMD ≤ (−3) (very strong hits). Blue star data points indicate the three hits characterized further in Figures 3, 4, 5, and 6. (D) Plate layout for the ON TARGETplus validation screen. (E) Replicate normalized TGN retrieval ratios for the ON TARGETplus smartpool validation screen (360 pools). The colors used are as in (C), with additional magenta data points indicating siRNA pools with TGN retrieval ratio SSMD between (−2) and (−3) (strong hits). (F) Table listing the 88 genes that were very strong or strong hits in the validation screen. Red-bordered cells indicate kinase and phosphatase genes, green-bordered cells contain genes encoding membrane proteins, and blue-bordered cells contain genes encoding cytoskeletal proteins. More information about these genes is provided in Table S4.
Figure 3
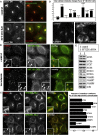
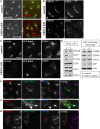
Characterization of SFT2D2 Function in Endosome-to-Golgi Retrieval (A) Primary screen anti-CD8 antibody-uptake images for control cells (top) and SFT2D2 KD cells (bottom). (B) Cells stably expressing SFT2D2-Myc show localization of SFT2D2 to perinuclear membranes and endosomes, including VPS35-positive endosomes (arrowheads in inset). Treatment with nocodazole disperses the endosomes and even more clearly shows colocalization of SFT2D2-Myc and VPS35 (arrowheads in inset). (C) Cells stably expressing SFT2D2-Myc were costained with antibodies against various post-Golgi SNARE proteins. Colocalization was quantified by Pearson’s correlation coefficient (right-hand graph) and indicates very extensive colocalization with STX6, STX7, and VAMP8. The image panels illustrate colocalization of SFT2D2-Myc and STX5 at the Golgi (top, arrowhead) but much more extensive colocalization of SFT2D2-Myc and STX6 (bottom, arrowheads). In (B) and (C), the white dashed box delineates the area magnified in the insets. (D) SNARE protein staining was compared for control and SFT2D2 KD HeLa cells and quantified. The graph shows the change in cellular intensity measured for each post-Golgi SNARE investigated. Images illustrate the increased cellular intensity of VAMP3 in SFT2D2 KD cells. (E) Control and SFT2D2 KD HeLa cell lysates were separated by LDS-PAGE and blotted for the indicated SNARE proteins or actin. Scale bars in (A)–(D), 20 μm, except insets in (B), 1 μm, and in (C), 2 μm. Quantitation in (C) and (D) was done using automated microscopy (see Experimental Procedures); error bars indicate SD of two separate multicell experiments. Average Pearson’s correlation coefficients measured were often identical in repeated experiments.
Figure 4
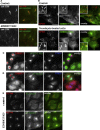
Localization and Characterization of ZDHHC5 in HeLa Cells (A) Primary screen anti-CD8 antibody-uptake images for control cells (top) and ZDHHC5 KD cells (bottom). (B) Cells stably expressing ZDHHC5-Myc show localization of ZDHHC5 to the plasma membrane and to intracellular tubules and vesicles. Some colocalization between ZDHHC5 and retromer VPS35 is observed (arrowheads in inset). Following nocodazole treatment endosomes are dispersed and some are labeled with ZDHHC5 and VPS35 (arrowheads in inset). (C) Control and ZDHHC5 siRNA-treated SFT2D2-Myc cells were mixed and stained for ZDHHC5, Myc, and VPS35. KD cells are marked by an asterisk. (D) Control and ZDHHC5 siRNA-treated HeLa cells were mixed and stained for ZDHHC5, α5-integrin, and VPS35. KD cells are indicated with an asterisk. (E) Control (top) and ZDHHC5 KD (bottom) HeLa cells were fixed and stained for TGN46 and β1-integrin. Scale bars in (A)–(E), 20 μm, except inset in (B), 2 μm.
Figure 5
GRINA Knockdown or Overexpression Inhibits Endosome-to-Golgi Retrieval (A) Primary screen anti-CD8 antibody uptake images for control cells (top) and GRINA KD cells (bottom). (B) Control (top) and GRINA KD (bottom) HeLa cells were stained for Golgi marker GM130 and Golgi glycoprotein-1 (GLG1). (C) Cells stably expressing GFP-Rab6 were treated with GRINA siRNA (bottom) and compared to control cells (top) upon staining for TGN46 and VPS35. (D) Quantitation by western blotting of lysates from control and GRINA-silenced HeLa cells and GFP-Rab6 cells. (E–G) HeLa cells were transiently transfected with GRINA-Myc for 24 hr before fixing and staining. (E and F) GRINA-Myc colocalizes with both TGN46 and VPS35. In this example, GRINA-Myc expression (in the cell marked by an asterisk in E) reduced the cell’s TGN46 expression compared to surrounding untransfected cells and caused enlargement of VPS35-positive endosomes. The area in (E) magnified in (F) is indicated by a dashed line. (G) GRINA-Myc expression also perturbs CIMPR localization. In this example, GRINA-Myc transfection (in the cell marked by an asterisk) caused CIMPR to localize to round vesicular structures positive for GRINA-Myc, some of which appeared larger than regular endosomes, whereas TGN46 staining was almost absent in the transfected cell. Scale bars in (A)–(C), (E), and (G), 20 μm, and in (F), 10 μm.
Figure 6
ZDHHC5, SFT2D2, or GRINA Depletion Affect Levels of Endosome-to-Golgi Cargo Proteins (A) Control HeLa cells or cells transfected with the indicated siRNAs were treated for 3 hr with cycloheximide, lysed, and incubated with agarose-bound wheat germ agglutinin to capture glycosylated membrane proteins. Total cell lysates (left) and lectin pull-down samples (right) were assayed by western blotting. The experiment was repeated three times, and representative data are shown. (B–E) Quantitative analysis of SFT2D2, ZDHHC5, and GRINA KD cells immunofluorescence using automated microscopy (see Experimental Procedures). (B) Representative images showing VPS35, TGN46, and CIMPR staining. Scale bar, 50 μm. (C) Quantitative analysis of CIMPR intensity at the Golgi indicates a significant increase in SFT2D2, ZDHHC5, or GRINA KD cells. (D) SFT2D2, ZDHHC5, and GRINA KD increase the Pearson’s correlation coefficient for colocalization between CIMPR and retromer proteins VPS35 or SNX1 while decreasing the correlation between CIMPR and Golgi matrix protein GM130. In some cases, identical correlation coefficients were measured in the replicate experiments. (E) Quantitation of the TGN46 and GLG1 intensity in the three types of KD cells. In (C)–(E), error bars indicate the SD of the replicate measurements. ∗p < 0.05; ∗∗p < 0.01.
Figure 7
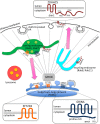
Model Schematic depiction of the localization and topology of the three multipass membrane proteins identified as regulators of endosome-to-Golgi retrieval. Features such as the YxxΦ motifs in ZDHHC5 are indicated.
Similar articles
- Retromer and the cation-independent mannose 6-phosphate receptor-Time for a trial separation?
Seaman MNJ. Seaman MNJ. Traffic. 2018 Feb;19(2):150-152. doi: 10.1111/tra.12542. Epub 2017 Dec 21. Traffic. 2018. PMID: 29135085 Free PMC article. Review. - EHD1 interacts with retromer to stabilize SNX1 tubules and facilitate endosome-to-Golgi retrieval.
Gokool S, Tattersall D, Seaman MNJ. Gokool S, et al. Traffic. 2007 Dec;8(12):1873-1886. doi: 10.1111/j.1600-0854.2007.00652.x. Epub 2007 Oct 7. Traffic. 2007. PMID: 17868075 - Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval.
Seaman MN. Seaman MN. J Cell Sci. 2007 Jul 15;120(Pt 14):2378-89. doi: 10.1242/jcs.009654. J Cell Sci. 2007. PMID: 17606993 - Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer.
Seaman MN. Seaman MN. J Cell Biol. 2004 Apr;165(1):111-22. doi: 10.1083/jcb.200312034. J Cell Biol. 2004. PMID: 15078902 Free PMC article. - Retromer-mediated endosomal protein sorting: The role of unstructured domains.
Mukadam AS, Seaman MN. Mukadam AS, et al. FEBS Lett. 2015 Sep 14;589(19 Pt A):2620-6. doi: 10.1016/j.febslet.2015.05.052. Epub 2015 Jun 10. FEBS Lett. 2015. PMID: 26072290 Review.
Cited by
- Retromer has a selective function in cargo sorting via endosome transport carriers.
Cui Y, Carosi JM, Yang Z, Ariotti N, Kerr MC, Parton RG, Sargeant TJ, Teasdale RD. Cui Y, et al. J Cell Biol. 2019 Feb 4;218(2):615-631. doi: 10.1083/jcb.201806153. Epub 2018 Dec 17. J Cell Biol. 2019. PMID: 30559172 Free PMC article. - A CRISPR Screen Identifies LAPTM4A and TM9SF Proteins as Glycolipid-Regulating Factors.
Yamaji T, Sekizuka T, Tachida Y, Sakuma C, Morimoto K, Kuroda M, Hanada K. Yamaji T, et al. iScience. 2019 Jan 25;11:409-424. doi: 10.1016/j.isci.2018.12.039. Epub 2019 Jan 3. iScience. 2019. PMID: 30660999 Free PMC article. - Retromer and the cation-independent mannose 6-phosphate receptor-Time for a trial separation?
Seaman MNJ. Seaman MNJ. Traffic. 2018 Feb;19(2):150-152. doi: 10.1111/tra.12542. Epub 2017 Dec 21. Traffic. 2018. PMID: 29135085 Free PMC article. Review. - PTPN23 binds the dynein adaptor BICD1 and is required for endocytic sorting of neurotrophin receptors.
Budzinska MI, Villarroel-Campos D, Golding M, Weston A, Collinson L, Snijders AP, Schiavo G. Budzinska MI, et al. J Cell Sci. 2020 Mar 30;133(6):jcs242412. doi: 10.1242/jcs.242412. J Cell Sci. 2020. PMID: 32079660 Free PMC article. - S-acylation of NLRP3 provides a nigericin sensitive gating mechanism that controls access to the Golgi.
Williams DM, Peden AA. Williams DM, et al. Elife. 2024 Sep 12;13:RP94302. doi: 10.7554/eLife.94302. Elife. 2024. PMID: 39263961 Free PMC article.
References
- Ahn J., Febbraio M., Silverstein R.L. A novel isoform of human Golgi complex-localized glycoprotein-1 (also known as E-selectin ligand-1, MG-160 and cysteine-rich fibroblast growth factor receptor) targets differential subcellular localization. J. Cell Sci. 2005;118:1725–1731. - PubMed
- Banfield D.K., Lewis M.J., Pelham H.R. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. - PubMed
- Belenkaya T.Y., Wu Y., Tang X., Zhou B., Cheng L., Sharma Y.V., Yan D., Selva E.M., Lin X. The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-Golgi network. Dev. Cell. 2008;14:120–131. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials