Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation - PubMed (original) (raw)
Review
Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation
Matthew R Bauerle et al. J Biol Chem. 2015.
Abstract
Radical S-adenosylmethionine (SAM) enzymes use the oxidizing power of a 5'-deoxyadenosyl 5'-radical to initiate an amazing array of transformations, usually through the abstraction of a target substrate hydrogen atom. A common reaction of radical SAM (RS) enzymes is the methylation of unactivated carbon or phosphorous atoms found in numerous primary and secondary metabolites, as well as in proteins, sugars, lipids, and RNA. However, neither the chemical mechanisms by which these unactivated atoms obtain methyl groups nor the actual methyl donors are conserved. In fact, RS methylases have been grouped into three classes based on protein architecture, cofactor requirement, and predicted mechanism of catalysis. Class A methylases use two cysteine residues to methylate sp(2)-hybridized carbon centers. Class B methylases require a cobalamin cofactor to methylate both sp(2)-hybridized and sp(3)-hybridized carbon centers as well as phosphinate phosphorous atoms. Class C methylases share significant sequence homology with the RS enzyme, HemN, and may bind two SAM molecules simultaneously to methylate sp(2)-hybridized carbon centers. Lastly, we describe a new class of recently discovered RS methylases. These Class D methylases, unlike Class A, B, and C enzymes, which use SAM as the source of the donated methyl carbon, are proposed to methylate sp(2)-hybridized carbon centers using methylenetetrahydrofolate as the source of the appended methyl carbon.
Keywords: Antibiotics; Cyclopropanation; Iron-Sulfur Protein; Methylation; Methylcobalamin; Methylenetetrahydrofolate; Radical; Ribosomal Ribonucleic Acid (rRNA) (Ribosomal RNA); S-Adenosylmethionine (SAM).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
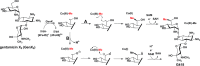
SCHEME 1.
Proposed mechanism for the RlmN-catalyzed reaction.
SCHEME 2.
Proposed mechanism for the GenK-catalyzed reaction.
SCHEME 3.
Reactions catalyzed by two class C RS methylases, YtkT (A) and Jaw5 (B). Two mechanisms for Jaw5 are shown utilizing a methylene radical (i) and an ylide (ii).
Similar articles
- Radical-mediated enzymatic methylation: a tale of two SAMS.
Zhang Q, van der Donk WA, Liu W. Zhang Q, et al. Acc Chem Res. 2012 Apr 17;45(4):555-64. doi: 10.1021/ar200202c. Epub 2011 Nov 18. Acc Chem Res. 2012. PMID: 22097883 Free PMC article. Review. - Surprise! A hidden B12 cofactor catalyzes a radical methylation.
Jarrett JT. Jarrett JT. J Biol Chem. 2019 Aug 2;294(31):11726-11727. doi: 10.1074/jbc.H119.009976. J Biol Chem. 2019. PMID: 31375551 Free PMC article. - Introduction to the thematic minireview series on radical S-adenosylmethionine (SAM) enzymes.
Banerjee R. Banerjee R. J Biol Chem. 2015 Feb 13;290(7):3962-3. doi: 10.1074/jbc.R114.630251. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477525 Free PMC article. Review. - The biosynthesis of thiol- and thioether-containing cofactors and secondary metabolites catalyzed by radical S-adenosylmethionine enzymes.
Jarrett JT. Jarrett JT. J Biol Chem. 2015 Feb 13;290(7):3972-9. doi: 10.1074/jbc.R114.599308. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477512 Free PMC article. Review. - Radical SAM-mediated methylation reactions.
Fujimori DG. Fujimori DG. Curr Opin Chem Biol. 2013 Aug;17(4):597-604. doi: 10.1016/j.cbpa.2013.05.032. Epub 2013 Jul 5. Curr Opin Chem Biol. 2013. PMID: 23835516 Free PMC article. Review.
Cited by
- Stereochemical course of cobalamin-dependent radical SAM methylation by TokK and ThnK.
Lichstrahl MS, Townsend CA, Sinner EK. Lichstrahl MS, et al. RSC Chem Biol. 2022 Jun 6;3(8):1028-1034. doi: 10.1039/d2cb00113f. eCollection 2022 Aug 3. RSC Chem Biol. 2022. PMID: 36042702 Free PMC article. - Anaerobic benzene oxidation in Geotalea daltonii involves activation by methylation and is regulated by the transition state regulator AbrB.
Bullows JE, Kanak A, Shedrick L, Kiessling C, Aklujkar M, Kostka J, Chin K-J. Bullows JE, et al. Appl Environ Microbiol. 2024 Oct 23;90(10):e0085624. doi: 10.1128/aem.00856-24. Epub 2024 Sep 17. Appl Environ Microbiol. 2024. PMID: 39287397 Free PMC article. - Consecutive radical S-adenosylmethionine methylations form the ethyl side chain in thienamycin biosynthesis.
Marous DR, Lloyd EP, Buller AR, Moshos KA, Grove TL, Blaszczyk AJ, Booker SJ, Townsend CA. Marous DR, et al. Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10354-8. doi: 10.1073/pnas.1508615112. Epub 2015 Aug 3. Proc Natl Acad Sci U S A. 2015. PMID: 26240322 Free PMC article. - Late-Stage C(sp 3)-H Methylation of Drug Molecules.
Mao E, MacMillan DWC. Mao E, et al. J Am Chem Soc. 2023 Feb 8;145(5):2787-2793. doi: 10.1021/jacs.2c13396. Epub 2023 Jan 25. J Am Chem Soc. 2023. PMID: 36696091 Free PMC article. - Radical SAM-dependent formation of a nitrogenase cofactor core on NifB.
Liu YA, Quechol R, Solomon JB, Lee CC, Ribbe MW, Hu Y, Hedman B, Hodgson KO. Liu YA, et al. J Inorg Biochem. 2022 Aug;233:111837. doi: 10.1016/j.jinorgbio.2022.111837. Epub 2022 Apr 20. J Inorg Biochem. 2022. PMID: 35550498 Free PMC article. Review.
References
- Takusagawa F., Fujioka M., Spies A., Schowen R. L. (1998) S-Adenosylmethionine (AdoMet)-dependent methyltransferases. in Comprehensive Biological Catalysis (Sinnott M. ed), pp. 1–30, Academic Press, New York
- Markham G. D. (2010) S-Adenosylmethionine. Encyclopedia of Life Sciences 10.1002/9780470015902.a0000662.pub2 - DOI
- Cantoni G. L. (1953) S-Adenosylmethionine: a new intermediate formed enzymatically from l-methionine and adenosine triphosphate. J. Biol. Chem. 204, 403–416 - PubMed
- Woodard R. W., Tsai M.-D., Floss H. G., Crooks P. A., Coward J. K. (1980) Sterochemical course of the transmethylation catalyzed by catechol O-methyltransferase. J. Biol. Chem. 255, 9124–9127 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources