Spore photoproduct lyase: the known, the controversial, and the unknown - PubMed (original) (raw)
Review
Spore photoproduct lyase: the known, the controversial, and the unknown
Linlin Yang et al. J Biol Chem. 2015.
Abstract
Spore photoproduct lyase (SPL) repairs 5-thyminyl-5,6-dihydrothymine, a thymine dimer that is also called the spore photoproduct (SP), in germinating endospores. SPL is a radical S-adenosylmethionine (SAM) enzyme, utilizing the 5'-deoxyadenosyl radical generated by SAM reductive cleavage reaction to revert SP to two thymine residues. Here we review the current progress in SPL mechanistic studies. Protein radicals are known to be involved in SPL catalysis; however, how these radicals are quenched to close the catalytic cycle is under debate.
Keywords: DNA Repair; DNA-Protein Interaction; Enzyme Catalysis; Enzyme Mechanism; Radiation Biology; Radical; Redox.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
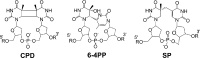
FIGURE 1.
The chemical structures of naturally occurring thymine photodimers.
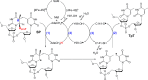
FIGURE 2.
The hypothesized reaction mechanism for SPL (the residues are numbered according to the protein sequence in B. subtilis SPL). This mechanism implies that SPL uses a minimum of four hydrogen atom transfer processes (labeled in blue numbers) in each catalytic cycle. The first two HAT processes are well established, and the last two HAT processes are under debate. SAM is shown to be regenerated at the end of the catalytic cycle, which is also controversial. (The figure is modified with permission from Ref. . Copyright (2013) American Chemical Society.)
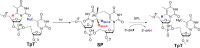
FIGURE 3.
Hydrogen atom migration during the SP formation and repair by SPL. A hydrogen atom migrates to the H6pro_S_ position of the formed SP, whereas the H6pro_R_ atom is abstracted to initiate the SP repair process. Therefore, the H6 atom of regenerated 5′-thymine after SPL repair is different from that before SP is formed. This observation provides the rationale for the previous tritium-labeling experiments (18, 26).
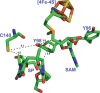
FIGURE 4.
The active site of G. thermodenitrificans (Gt) SPL in complex with SP and SAM. Cys-140(Gt), Tyr-96(Gt), and Tyr-98(Gt) equal to Cys-141(Bs), Tyr-97(Bs), and Tyr-99(Bs) in B. subtilis SPL, respectively. The distances (Å) between selected residues, SP, and SAM are indicated by the black numbers near the dashed lines (Protein Data Bank (PDB) code 4FHD).
Similar articles
- Mechanistic studies of the radical SAM enzyme spore photoproduct lyase (SPL).
Li L. Li L. Biochim Biophys Acta. 2012 Nov;1824(11):1264-77. doi: 10.1016/j.bbapap.2011.11.008. Epub 2011 Dec 8. Biochim Biophys Acta. 2012. PMID: 22197590 Free PMC article. Review. - Characterization of an active spore photoproduct lyase, a DNA repair enzyme in the radical S-adenosylmethionine superfamily.
Buis JM, Cheek J, Kalliri E, Broderick JB. Buis JM, et al. J Biol Chem. 2006 Sep 8;281(36):25994-6003. doi: 10.1074/jbc.M603931200. Epub 2006 Jul 7. J Biol Chem. 2006. PMID: 16829680 - Probing the reaction mechanism of spore photoproduct lyase (SPL) via diastereoselectively labeled dinucleotide SP TpT substrates.
Yang L, Lin G, Liu D, Dria KJ, Telser J, Li L. Yang L, et al. J Am Chem Soc. 2011 Jul 13;133(27):10434-47. doi: 10.1021/ja110196d. Epub 2011 Jun 14. J Am Chem Soc. 2011. PMID: 21671623 Free PMC article. - The enzyme-mediated direct reversal of a dithymine photoproduct in germinating endospores.
Yang L, Li L. Yang L, et al. Int J Mol Sci. 2013 Jun 25;14(7):13137-53. doi: 10.3390/ijms140713137. Int J Mol Sci. 2013. PMID: 23799365 Free PMC article. - DNA Repair by the Radical SAM Enzyme Spore Photoproduct Lyase: From Biochemistry to Structural Investigations.
Berteau O, Benjdia A. Berteau O, et al. Photochem Photobiol. 2017 Jan;93(1):67-77. doi: 10.1111/php.12702. Epub 2017 Jan 27. Photochem Photobiol. 2017. PMID: 28027411 Review.
Cited by
- The small acid-soluble proteins of Clostridioides difficile regulate sporulation in a SpoIVB2-dependent manner.
Nerber HN, Baloh M, Brehm JN, Sorg JA. Nerber HN, et al. PLoS Pathog. 2024 Aug 30;20(8):e1012507. doi: 10.1371/journal.ppat.1012507. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39213448 Free PMC article. - Insights into the Activity Change of Spore Photoproduct Lyase Induced by Mutations at a Peripheral Glycine Residue.
Yang L, Li L. Yang L, et al. Front Chem. 2017 Mar 28;5:14. doi: 10.3389/fchem.2017.00014. eCollection 2017. Front Chem. 2017. PMID: 28401144 Free PMC article. - Catalytic Promiscuity of the Radical S-adenosyl-L-methionine Enzyme NosL.
Ding W, Ji X, Li Y, Zhang Q. Ding W, et al. Front Chem. 2016 Jun 22;4:27. doi: 10.3389/fchem.2016.00027. eCollection 2016. Front Chem. 2016. PMID: 27446906 Free PMC article. Review. - Kinetic Isotope Effects and Hydrogen/Deuterium Exchange Reveal Large Conformational Changes During the Catalysis of the Clostridium acetobutylicum Spore Photoproduct Lyase.
Yang L, Adhikari J, Gross ML, Li L. Yang L, et al. Photochem Photobiol. 2017 Jan;93(1):331-342. doi: 10.1111/php.12697. Epub 2017 Jan 30. Photochem Photobiol. 2017. PMID: 27992649 Free PMC article. - Small Prokaryotic DNA-Binding Proteins Protect Genome Integrity throughout the Life Cycle.
Molan K, Žgur Bertok D. Molan K, et al. Int J Mol Sci. 2022 Apr 4;23(7):4008. doi: 10.3390/ijms23074008. Int J Mol Sci. 2022. PMID: 35409369 Free PMC article. Review.
References
- Cadet J., Vigny P. (1990) in: Bioorganic Photochemistry, Vol. 1, Photochemistry and the Nucleic Acids (Morrison H., ed) pp. 1–272, John Wiley & Sons, Inc., New York
- Setlow P. (2001) Resistance of spores of Bacillus species to ultraviolet light. Environ. Mol. Mutagen. 38, 97–104 - PubMed
- Desnous C., Guillaume D., Clivio P. (2010) Spore photoproduct: a key to bacterial eternal life. Chem. Rev. 110, 1213–1232 - PubMed
- Setlow P. (1988) Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Annu. Rev. Microbiol. 42, 319–338 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources