Posttranslational modification of autophagy-related proteins in macroautophagy - PubMed (original) (raw)
Review
Posttranslational modification of autophagy-related proteins in macroautophagy
Yangchun Xie et al. Autophagy. 2015.
Abstract
Macroautophagy is an intracellular catabolic process involved in the formation of multiple membrane structures ranging from phagophores to autophagosomes and autolysosomes. Dysfunction of macroautophagy is implicated in both physiological and pathological conditions. To date, 38 autophagy-related (ATG) genes have been identified as controlling these complicated membrane dynamics during macroautophagy in yeast; approximately half of these genes are clearly conserved up to human, and there are additional genes whose products function in autophagy in higher eukaryotes that are not found in yeast. The function of the ATG proteins, in particular their ability to interact with a number of macroautophagic regulators, is modulated by posttranslational modifications (PTMs) such as phosphorylation, glycosylation, ubiquitination, acetylation, lipidation, and proteolysis. In this review, we summarize our current knowledge of the role of ATG protein PTMs and their functional relevance in macroautophagy. Unraveling how these PTMs regulate ATG protein function during macroautophagy will not only reveal fundamental mechanistic insights into the regulatory process, but also provide new therapeutic targets for the treatment of autophagy-associated diseases.
Keywords: AMPK, AMP-activated protein kinase; ATG, autophagy-related; MTORC1, mechanistic target of rapamycin complex 1; PE, phosphatidylethanolamine; PTM, posttranslational modification; Ub, ubiquitin; Ubl, ubiquitin like; autophagy; autophagy-related proteins; posttranslational modification.
Figures
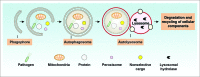
Figure 1.
Overview of the autophagy process. Autophagy is a lysosome-mediated degradation and recycling pathway that involves the formation of multiple membrane structures ranging from phagophores to autophagosomes and autolysosomes.
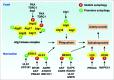
Figure 2.
Regulation of Atg/ATG proteins by phosphorylation. In yeast, the initiation of autophagy involves the inhibition of TORC1- and PKA-mediated phosphorylation of Atg1 and Atg13 under starvation conditions. Atg1 is then activated via autophosphorylation, which is responsible for Atg9 localization to the phagophore assembly site. In addition, phosphorylation of Atg29 stimulates autophagy by activating the Atg17-Atg31-Atg29 complex, which is required for subsequent interaction with Atg11. In mammalian cells, phosphorylation of ULK1 and BECN1 by the indicated kinases has dual roles in the regulation of the autophagic response. Phosphorylation of LC3 and ATG5 by PRKAC and MAPK14, respectively, inhibits autophagy. In addition, ULK1- and ATG101-mediated ATG13 phosphorylation promotes selective autophagy.
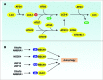
Figure 3.
Regulation of ATG proteins by ubiquitination. (A) Two Ubl conjugation systems in mammalian cells. (B) Ubiquitination or deubiquitination of BECN1 and ULK1 regulate the autophagic response.
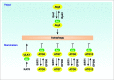
Figure 4.
Regulation of Atg/ATG proteins by acetylation. In yeast, acetylation of Atg3 by Esa1 increases autophagy, whereas deacetylation of Atg3 by Rpd3 decreases autophagy. In mammalian cells, acetylation of ATGs (ATG5, ATG7, ATG8, and ATG12) by EP300 decreases autophagy, whereas deacetylation of these ATGs by SIRT1 increases autophagy. In addition, KAT5-mediated ULK1 acetylation promotes autophagy.
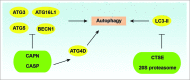
Figure 5.
Regulation of ATG proteins by proteolysis. CAPN/calpain and CASP can inhibit autophagy by degrading or cleaving ATGs such as ATG3, ATG5, ATG16L1, and BECN1. In contrast, CASP-mediated ATG4D cleavage promotes autophagy. In addition, CTSE/cathepsin E and the 20S proteasome are involved in the degradation of the phagophore and autophagosome membrane component LC3-II.
Similar articles
- Deacetylation of ATG7 drives the induction of macroautophagy and LC3-associated microautophagy.
Xu Y, Qian C, Wang Q, Song L, He Z, Liu W, Wan W. Xu Y, et al. Autophagy. 2024 May;20(5):1134-1146. doi: 10.1080/15548627.2023.2287932. Epub 2023 Nov 28. Autophagy. 2024. PMID: 37999993 Free PMC article. - Chemical Biology of Autophagy-Related Proteins With Posttranslational Modifications: From Chemical Synthesis to Biological Applications.
Luo Y, Jiang C, Yu L, Yang A. Luo Y, et al. Front Chem. 2020 Apr 3;8:233. doi: 10.3389/fchem.2020.00233. eCollection 2020. Front Chem. 2020. PMID: 32309274 Free PMC article. Review. - Assessment of Posttranslational Modifications of ATG proteins.
Xie Y, Kang R, Tang D. Xie Y, et al. Methods Enzymol. 2017;587:171-188. doi: 10.1016/bs.mie.2016.09.057. Epub 2016 Nov 11. Methods Enzymol. 2017. PMID: 28253954 - THANATOS: an integrative data resource of proteins and post-translational modifications in the regulation of autophagy.
Deng W, Ma L, Zhang Y, Zhou J, Wang Y, Liu Z, Xue Y. Deng W, et al. Autophagy. 2018;14(2):296-310. doi: 10.1080/15548627.2017.1402990. Autophagy. 2018. PMID: 29157087 Free PMC article. - Fine-tuning autophagy: from transcriptional to posttranslational regulation.
Botti-Millet J, Nascimbeni AC, Dupont N, Morel E, Codogno P. Botti-Millet J, et al. Am J Physiol Cell Physiol. 2016 Sep 1;311(3):C351-62. doi: 10.1152/ajpcell.00129.2016. Epub 2016 Jun 22. Am J Physiol Cell Physiol. 2016. PMID: 27335173 Review.
Cited by
- Cuproptosis: Mechanisms, biological significance, and advances in disease treatment-A systematic review.
Pan C, Ji Z, Wang Q, Zhang Z, Wang Z, Li C, Lu S, Ge P. Pan C, et al. CNS Neurosci Ther. 2024 Sep;30(9):e70039. doi: 10.1111/cns.70039. CNS Neurosci Ther. 2024. PMID: 39267265 Free PMC article. Review. - Upregulation of KCNQ1OT1 promotes resistance to stereotactic body radiotherapy in lung adenocarcinoma by inducing ATG5/ATG12-mediated autophagy via miR-372-3p.
He H, Song X, Yang Z, Mao Y, Zhang K, Wang Y, Su B, Li Q, Chen H, Li Y. He H, et al. Cell Death Dis. 2020 Oct 20;11(10):883. doi: 10.1038/s41419-020-03083-8. Cell Death Dis. 2020. PMID: 33082306 Free PMC article. - WERAM: a database of writers, erasers and readers of histone acetylation and methylation in eukaryotes.
Xu Y, Zhang S, Lin S, Guo Y, Deng W, Zhang Y, Xue Y. Xu Y, et al. Nucleic Acids Res. 2017 Jan 4;45(D1):D264-D270. doi: 10.1093/nar/gkw1011. Epub 2016 Oct 26. Nucleic Acids Res. 2017. PMID: 27789692 Free PMC article. - Autophagy: An Essential Degradation Program for Cellular Homeostasis and Life.
Chun Y, Kim J. Chun Y, et al. Cells. 2018 Dec 19;7(12):278. doi: 10.3390/cells7120278. Cells. 2018. PMID: 30572663 Free PMC article. Review. - Understanding and exploiting the roles of autophagy in plants through multi-omics approaches.
Liu F, Marshall RS, Li F. Liu F, et al. Plant Sci. 2018 Sep;274:146-152. doi: 10.1016/j.plantsci.2018.05.009. Epub 2018 May 22. Plant Sci. 2018. PMID: 30080598 Free PMC article. Review.
References
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814-22; PMID:20811353; http://dx.doi.org/10.1038/ncb0910-814 - DOI - PMC - PubMed
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/10.4161/auto.19496 - DOI - PMC - PubMed
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313-26; PMID:20144757; http://dx.doi.org/10.1016/j.cell.2010.01.028 - DOI - PMC - PubMed
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469:323-35; PMID:21248839; http://dx.doi.org/10.1038/nature09782 - DOI - PMC - PubMed
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8:741-52; PMID:17717517; http://dx.doi.org/10.1038/nrm2239 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous