Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism - PubMed (original) (raw)
Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism
Wen-Chuan Shen et al. Autophagy. 2015.
Abstract
OPTN (optineurin) is an autophagy receptor and mutations in the OPTN gene result in familial glaucoma (E50K) and amyotrophic lateral sclerosis (ALS) (E478G). However, the mechanisms through which mutant OPTN leads to human diseases remain to be characterized. Here, we demonstrated that OPTN colocalized with inclusion bodies (IBs) formed by mutant HTT/huntingtin protein (mHTT) in R6/2 transgenic mice and IBs formed by 81QNmHTT (nuclear form), 109QmHTT (cytoplasmic form) or the truncated form of TARDBP/TDP-43 (TARDBP(ND251)) in Neuro2A cells. This colocalization required the ubiquitin (Ub)-binding domain (UbBD, amino acids 424 to 511) of OPTN. Overexpression of wild-type (WT) OPTN decreased IBs through K63-linked polyubiquitin-mediated autophagy. E50K or 210 to 410Δ (with amino acids 210 to 410 deleted) whose mutation or deletion was outside the UbBD decreased the IBs formed by 109QmHTT or TARDBP(ND251), as was the case with WT OPTN. In contrast, UbBD mutants, including E478G, D474N, UbBDΔ, 411 to 520Δ and 210 to 520Δ, increased accumulation of IBs. UbBD mutants (E478G, UbBDΔ) retained a substantial ability to interact with WT OPTN, and were found to colocalize with polyubiquitinated IBs, which might occur indirectly through their WT partner in a WT-mutant complex. They decreased autophagic flux evidenced by alteration in LC3 level and turnover and in the number of LC3-positive puncta under stresses like starvation or formation of IBs. UbBD mutants exhibited a weakened interaction with MYO6 (myosin VI) and TOM1 (target of myb1 homolog [chicken]), important for autophagosome maturation, in cells or sorted 109QmHtt IBs. Taken together, our data indicated that UbBD mutants acted as dominant-negative traps through the formation of WT-mutant hybrid complexes to compromise the maturation of autophagosomes, which in turn interfered with OPTN-mediated autophagy and clearance of IBs.
Keywords: ALS, amyotrophic lateral sclerosis; Ab, antibody; BafA1, bafilomycin A1; CCD, coiled-coil domain; Ef, FRET efficiency; FT, filter-trap assay; HD, Huntington disease; IBs, inclusion bodies; IP, immunoprecipitation; K48, lysine 48; K63, lysine 63; LIR, LC3-interacting region; MYO6, myosin VI; OPTN, optineurin; PBS, phosphate-buffered saline; PFA, paraformaldehyde; TARDBP/TDP-43; TARDBP/TDP-43, TAR DNA-binding protein; TBK1, TANK-binding kinase 1; TUBA, alpha tubulin; UPS, ubiquitin-proteasome system; Ub, ubiquitin B/C/D; UbBD, ubiquitin-binding domain; WB, western blot; WT, wild type; autophagy; dominant-negative; huntingtin; mHTT, mutant huntingtin; optineurin.
Figures
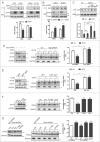
Figure 1.
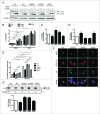
Colocalization of OPTN with IBs through UbBD. (A) Immunofluorescent confocal images of cortical and striatal sections of the 12-wk-old R6/2 transgenic mice stained with EM48 (mHTT), anti-ubiquitin (Ub) and anti-OPTN antibodies (Abs). (B) Confocal images of Neuro2A cells expressing GFP-81QNmHTT or GFP-109QmHTT stained with anti-Ub or anti-OPTN Abs. Arrows show IBs with signals. (C) Images of Neuro2A cells expressing eGFP-TARDBPND251 (ND251) stained with anti-OPTN Ab. (D) Western blot of the sorted GFP, GFP-positive 25QHTT, 109QmHTT or ND251 particles from IBs (P1) or non-IB (P2) fraction of the Neuro2A cells probed with anti-OPTN, EM48 or GFP Abs. Endogenous OPTN was detected only in 109QmHTT and ND251 IBs. (E) Schematic illustration of wild-type (WT) and truncated or mutated OPTN constructs. (F) Confocal images of Neuro2A cells co-overexpressing 109QmHTT and OPTN variants as indicated. (G) Quantitative graph of colocalization of the OPTN variants with 109QmHTT IBs. ***P < 0.001, ****P < 0.0001. Scale bar: 5 μm. At least 100 randomly chosen aggregates were calculated in triplicate experiments.
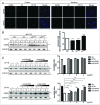
Figure 2.
OPTN reduces mHTT IBs through autophagy. (A) Filter trap (FT) assay for IBs of Neuro2A cells coexpressing vector or OPTN with 25QHTT (cytoplasmic), 25QNHTT (nuclear), 109QmHTT (cytoplasmic) or 81QNmHTT (nuclear). The corresponding western blotting (WB) showed equal expression of OPTN and loading (GAPDH). Quantitative graph (below) showing OPTN overexpression reduced both 109Q and 81QN IBs. (B) FT assay of Neuro2A cells with OPTN knockdown (KD) which increased IBs of both mHTT. TUBA, alpha-tubulin. (C) FT assay to show that OPTN-mediated reduction in the 109QmHTT IBs was blocked by 5 mM 3-methyladenine (3MA) or 2 μM bafilomycin A1 (BafA1), but not by 1 μM MG132. (D) Western blot analysis and quantification of the soluble (S, non-IB) and insoluble (I, IB) fractions of Neuro2A cells transfected with constructs as indicated. Please note knockdown of Atg5 by siRNA blocked the OPTN-mediated reduction in insoluble 109QmHTT (asterisks). (E) Similar experiments were conducted in MEF and _Atg5_-null MEF (_atg5_−/−) cells. (F) Western blot analysis and quantification showed that the LIR mutant, F178A or LIRΔ, had no significant effect on insolubility of mHTT compared with WT OPTN. (G) Functional rescue assays of WT and mutant OPTN in Neuro2A cells with knockdown of endogenous Optn by shRNA. The left subpanel indicates expression of WT and mutant OPTN. F178A or E478G failed to rescue the defect of OPTN knockdown as WT. All quantified data were collected from 3 independently performed experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001, ns: not significant.
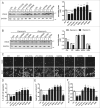
Figure 3.
OPTN-mediated 109QmHTT reduction occurs preferentially through polyK63Ub. (A) Confocal images of sections of cortex or striatum of 12-wk-old R6/2 mice stained with EM48, anti-OPTN, anti-UbK48- (UbK48) or anti-UbK63-specific (UbK63) Abs. Scale bar: 10 μm. (B) Western blot of Neuro2A cells coexpressing 109QmHTT, vector or OPTN plus the UbK48R or UbK63R mutant as indicated, and treated with 1 μM MG132 for 16 h. The reduction fold was calculated first with the insolubility index of the OPTN+ sample normalized with the corresponding OPTN- control. Then the Ub with OPTN or Ub mutant with OPTN sample was again normalized with Ctrl with OPTN (set as 1). The UbK63R mutant blocked the effect of OPTN. (C) Western blot of Neuro2A cells coexpressing 109QmHTT with vector (upper panel) or OPTN (lower panel), and treated with polyUbK48 or polyUbK63 peptide chain at 1, 2, or 4 μg/ml (left). The effect of OPTN was compromised by polyUbK63 more significantly than by polyUbK48. The corresponding expression of transfected protein for (B) and (C) is shown in Fig. S10A and S10B. Asterisks on the blots indicate the insoluble fractions. **P < 0.01, ***P < 0.001, ****P <0.0001, ns: not significant. All quantified data were collected from 3 independently performed experiments.
Figure 4.
Block of the OPTN-mediated decrease in 109QmHTT IBs by UbBD mutants. (A) Western blot of Neuro2A lysates coexpressing 109QmHTT with WT, truncated or mutated OPTN as indicated. UbBD mutants, 210 to 520Δ, 411 to 520Δ, UbBDΔ, E478G, D474N, increased insolubility index of 109QmHTT, unlike WT or E50K. (B) Western blot of cells coexpressing 109QmHTT with OPTN variant treated with or without 2 μM rapamycin for 24 h. The increased insolubility index by UbBD mutants was not rescued by rapamycin. Corresponding expression of transfected OPTN and mutants was shown in Fig. S10C for (A) and S10D for (B). Asterisks on the blots indicate the insoluble fractions. (C) Representative confocal images of Neuro2A cells coexpressing 109QmHTT and OPTN variants acquired by the ImageXpress Micro Imaging XL System. Scale bar: 20 μm. (D–F) Quantitative measurements of 109QmHTT IBs in Neuro2A images acquired in (C). *P < 0.05, ***P < 0.001, ****P < 0.0001, ns: not significant. All quantified data were collected from 3 independently performed experiments.
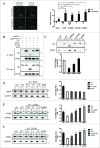
Figure 5.
UbBD mutants interacting with WT OPTN and acting as dominant-negative trap. (A) Images of Neuro2A cells coexpressing Cerulean-WT OPTN (Ce-WT) and Venus-WT OPTN (Ve-WT) with region of interest (ROI) (boxed) selected for acceptor photobleaching FRET study (Left). Quantification of FRET efficiency of Neuro2A cells coexpression of Ce-WT and Ve-WT or mutant was shown on the right. The same ROI before photobleaching (prebleach) served as Ctrl. (B) Interaction of tagRFP-WT OPTN with His-WT OPTN or mutants by immunoprecipitation with anti-tRFP antibody. Immunoblot was done with anti-His (for OPTN variants) and anti-OPTN antibodies. (C) Western blot and quantification of the level of WT OPTN and mutants associated with 109QmHTT IBs isolated from Neuro2A cells. The corresponding protein expression is shown in Figure S10E. (D, E, F) Competition assays in Neuro2A cells showing repression of WT OPTN function by E50K, UbBDΔ or E478G, respectively. The corresponding expression of transfected protein is shown in Fig. S5. Asterisks on the blots indicate the insoluble fractions. **P < 0.01, ***P < 0.001, ****P < 0.0001, ns: not significant. All quantified data were collected from 3 independently performed experiments.
Figure 6.
For figure legend, see page 695.Figure 6 (See previous page). Decreased autophagic flux by UbBD mutants. (A) Western blot and (B) corresponding quantification of LC3-II in Neuro2A cells expressing WT or mutant OPTN under fed (starvation-) or starvation (+) conditions, treated with DMSO (−) or BafA1, as indicated. The corresponding protein expression is shown in Fig. S10F. LC3-II expression was normalized against GAPDH. (C) Quantitative data on the effect of UbBD mutants on the fold change in LC3-II in response to starvation. The fold change was first calculated with the value of each sample in the starvation with DMSO group divided by corresponding sample in FED with DMSO group, which were then normalized with the Ctrl (set as 1). (D) Effect of UBbD mutants on LC3 turnover. Quantitative data were first calculated with the value of each sample in the starvation plus BafA1 group divided by corresponding sample in the starvation plus DMSO groups, which were then normalized with the Ctrl (set as 1). (E) Effect of the UbBD mutants on the number of GFP-LC3 dots from the images acquired by the ImageXpress Micro Imaging XL System (Fig. S7) with the indicated conditions on MCF7 cells stably expressing GFP-LC3. The corresponding protein expression was shown in Figure S10G. (F) Colocalization of WT and the mutant OPTN shown in confocal micrographs of Neuro2A cells co-overexpressing 109QmHTT, RFP-LC3 plus WT or mutant OPTN as indicated. Scale bar: 5 μm. (G) Western blot analysis of the RFP-LC3-II level in the isolated 109QmHTT IBs (P1) or soluble fraction (P2) from cells coexpressing WT or mutant OPTN as indicated (upper panel). Notably, LC3 was highly enriched in IB fraction. Quantitfication of the LC3-II level was shown in lower panel. Corresponding protein expression is shown in Fig. S10H. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. All quantified data were collected from 3 independently performed experiments.
Figure 7.
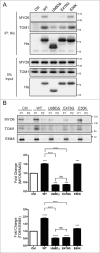
Reduced interaction between MYO6 and UbBD mutants. (A) MYO6 and TOM1 was pulled down from cells expressing His-WT or mutant OPTN as indicated with Ni-SepharoseTM beads. Cells were treated with 200 nM BafA1 for 16 h before harvest. Western blots were performed with antibodies as indicated in IP panel. (B) The level of MYO6 and TOM1 associated with the sorted 109QmHTT IBs from Neuro2A cells coexpressing WT or mutant OPTN. Corresponding protein expression is shown in Fig. S10E. ***P < 0.001, ****P < 0.0001, ns: not significant. All quantified data were collected from 3 independently performed experiments.
Figure 8.
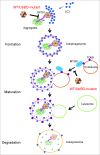
The schematic model of UbBD mutants as dominant-negative traps to interfere with autophagy-mediated IB degradation. UbBD-mutants interacted with the WT counterpart, and formed an WT-mutant hybrid oligomer or complex. Given their weaker affinity with IBs, MYO6, and TOM1, they decreased autophagy flux at both early and late stages and subsequent degradation of IBs.
Similar articles
- Defects in optineurin- and myosin VI-mediated cellular trafficking in amyotrophic lateral sclerosis.
Sundaramoorthy V, Walker AK, Tan V, Fifita JA, Mccann EP, Williams KL, Blair IP, Guillemin GJ, Farg MA, Atkin JD. Sundaramoorthy V, et al. Hum Mol Genet. 2015 Jul 1;24(13):3830-46. doi: 10.1093/hmg/ddv126. Epub 2015 Apr 9. Hum Mol Genet. 2015. PMID: 25859013 - Mutations of optineurin in amyotrophic lateral sclerosis.
Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Maruyama H, et al. Nature. 2010 May 13;465(7295):223-6. doi: 10.1038/nature08971. Epub 2010 Apr 28. Nature. 2010. PMID: 20428114 - A critical role of Hrd1 in the regulation of optineurin degradation and aggresome formation.
Mao J, Xia Q, Liu C, Ying Z, Wang H, Wang G. Mao J, et al. Hum Mol Genet. 2017 May 15;26(10):1877-1889. doi: 10.1093/hmg/ddx096. Hum Mol Genet. 2017. PMID: 28334804 - Altered Functions and Interactions of Glaucoma-Associated Mutants of Optineurin.
Swarup G, Sayyad Z. Swarup G, et al. Front Immunol. 2018 Jun 6;9:1287. doi: 10.3389/fimmu.2018.01287. eCollection 2018. Front Immunol. 2018. PMID: 29951055 Free PMC article. Review. - Optineurin: The autophagy connection.
Ying H, Yue BY. Ying H, et al. Exp Eye Res. 2016 Mar;144:73-80. doi: 10.1016/j.exer.2015.06.029. Epub 2015 Jul 2. Exp Eye Res. 2016. PMID: 26142952 Free PMC article. Review.
Cited by
- RNA Binding Proteins and the Pathogenesis of Frontotemporal Lobar Degeneration.
Hofmann JW, Seeley WW, Huang EJ. Hofmann JW, et al. Annu Rev Pathol. 2019 Jan 24;14:469-495. doi: 10.1146/annurev-pathmechdis-012418-012955. Epub 2018 Oct 24. Annu Rev Pathol. 2019. PMID: 30355151 Free PMC article. Review. - Autophagy in Neurodegenerative Diseases: A Hunter for Aggregates.
Park H, Kang JH, Lee S. Park H, et al. Int J Mol Sci. 2020 May 10;21(9):3369. doi: 10.3390/ijms21093369. Int J Mol Sci. 2020. PMID: 32397599 Free PMC article. Review. - The proteasome as a druggable target with multiple therapeutic potentialities: Cutting and non-cutting edges.
Tundo GR, Sbardella D, Santoro AM, Coletta A, Oddone F, Grasso G, Milardi D, Lacal PM, Marini S, Purrello R, Graziani G, Coletta M. Tundo GR, et al. Pharmacol Ther. 2020 Sep;213:107579. doi: 10.1016/j.pharmthera.2020.107579. Epub 2020 May 19. Pharmacol Ther. 2020. PMID: 32442437 Free PMC article. - Aberrant Stress Granule Dynamics and Aggrephagy in ALS Pathogenesis.
Zhang Y, Gu J, Sun Q. Zhang Y, et al. Cells. 2021 Aug 30;10(9):2247. doi: 10.3390/cells10092247. Cells. 2021. PMID: 34571896 Free PMC article. Review. - ALS Genetics: Gains, Losses, and Implications for Future Therapies.
Kim G, Gautier O, Tassoni-Tsuchida E, Ma XR, Gitler AD. Kim G, et al. Neuron. 2020 Dec 9;108(5):822-842. doi: 10.1016/j.neuron.2020.08.022. Epub 2020 Sep 14. Neuron. 2020. PMID: 32931756 Free PMC article. Review.
References
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nature medicine 2004; 10 Suppl:S10-7. - PubMed
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. . Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314:130-3. - PubMed
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. . TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochemical and biophysical research communications 2006; 351:602-11. - PubMed
- Ince PG, Highley JR, Kirby J, Wharton SB, Takahashi H, Strong MJ, et al. . Molecular pathology and genetic advances in amyotrophic lateral sclerosis: an emerging molecular pathway and the significance of glial pathology. Acta neuropathologica 2011; 122:657-71. - PubMed
- Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell 2010; 143:682-5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous