GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases - PubMed (original) (raw)
. 2015 Feb;33(2):187-197.
doi: 10.1038/nbt.3117. Epub 2014 Dec 16.
Shengdar Q Tsai # 1 2 3, Zongli Zheng # 1 2 3 4, Matthew Liebers 1 2, Ved V Topkar 1 2, Vishal Thapar 1 2, Nicolas Wyvekens 1 2, Cyd Khayter 1 2, A John Iafrate 1 2 3, Long P Le 1 2 3, Martin J Aryee 1 2 3, J Keith Joung 1 2 3
Affiliations
- PMID: 25513782
- PMCID: PMC4320685
- DOI: 10.1038/nbt.3117
GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases
Shengdar Q Tsai et al. Nat Biotechnol. 2015 Feb.
Abstract
CRISPR RNA-guided nucleases (RGNs) are widely used genome-editing reagents, but methods to delineate their genome-wide, off-target cleavage activities have been lacking. Here we describe an approach for global detection of DNA double-stranded breaks (DSBs) introduced by RGNs and potentially other nucleases. This method, called genome-wide, unbiased identification of DSBs enabled by sequencing (GUIDE-seq), relies on capture of double-stranded oligodeoxynucleotides into DSBs. Application of GUIDE-seq to 13 RGNs in two human cell lines revealed wide variability in RGN off-target activities and unappreciated characteristics of off-target sequences. The majority of identified sites were not detected by existing computational methods or chromatin immunoprecipitation sequencing (ChIP-seq). GUIDE-seq also identified RGN-independent genomic breakpoint 'hotspots'. Finally, GUIDE-seq revealed that truncated guide RNAs exhibit substantially reduced RGN-induced, off-target DSBs. Our experiments define the most rigorous framework for genome-wide identification of RGN off-target effects to date and provide a method for evaluating the safety of these nucleases before clinical use.
Figures
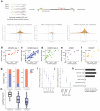
Figure 1. Design, optimization and application of the GUIDE-Seq method
(a) Schematic overview of the GUIDE-Seq method. (b) Optimization of dsODN integration into RGN-induced DSBs in human cells. Rates of integration for different modified oligonucleotides as measured by RFLP assay are shown. Control reactions were transfected with only the RGN-encoding plasmids (i.e., without dsODN). (c) Schematic illustrating how mapping of genomic sequence reads enabled identification of DSB position. (d) GUIDE-Seq-based identification of RGN-induced DSBs. Start sites of GUIDE-Seq reads mapped back to the genome enable localization of the DSB to within a few base pairs. Mapped reads for the on-target sites of the ten RGNs we assessed by GUIDE-Seq are shown. In all cases, the target site sequence is shown with the 20 bp protospacer sequence to the left and the PAM sequence to the right on the x-axis. Note how in all cases the highest peak falls within 3 to 4 bps of the 5′-edge of the NGG PAM sequence, the expected position of an RGN cleavage event. Equivalent maps for off-target sites are presented in Supplementary Fig. 2. (e) Numbers of previously known and novel off-target cleavage sites identified by GUIDE-Seq for the ten RGNs analyzed in this study. (f) Scatterplot of on-target site orthogonality to the human genome (y-axis) versus total number of off-target sites detected by GUIDE-Seq for the ten RGNs of this report. Orthogonality was calculated as the total number of sites in the human genome bearing 1 to 6 mismatches relative to the on-target site. (g) Scatterplot of on-target site GC content (y-axis) versus total number of off-target sites detected by GUIDE-Seq for the ten RGNs of this report. (h) Chromosome ideogram of CRISPR/Cas9 on- and off-target sites for the RGN that targets EMX1. Additional ideograms for the remaining RGNs can be found in Supplementary Fig. 3. (i) Genomic locations of off-target cleavage sites identified by GUIDE-Seq for the ten RGNs examined in this study.
Figure 2. Sequences of off-target sites identified by GUIDE-Seq for ten RGNs
For each RGN, the intended target sequence is shown in the top line with cleaved sites shown underneath and with mismatches to the on-target site shown and highlighted in color. GUIDE-Seq sequencing read counts are shown to the right of each site. The on-target site is marked with a green square and previously known off-target sites with a red diamond. Data is shown for RGNs targeting the following sites: (a) VEGFA site 1, (b) VEGFA site 2, (c) VEGFA site 3, (d) EMX1, (e) FANCF, (f) HEK293 site 1, (g) HEK293 site 2, (h) HEK293 site 3, (i) HEK293 site4, (j) RNF2. No off-target sites were found for the RGN targeted to the RNF2 site.
Figure 3. Analysis of RGN-induced off-target sequence characteristics
(a) Schematic overview of the AMP-based sequencing method used to confirm indel mutations at GUIDE-Seq cleavage sites is shown in the top half of the figure. Histogram plots of mapped indel mutations are shown for three RGN on-target sites. Deletions are shown above the X-axis whereas insertions are shown below. Boundaries of the overall target site (i.e., protospacer and PAM sequence) are shown with dotted lines and the boundary between the protospacer and PAM sequence is shown as a dotted line between the other two. RGN cleavage is predicted to occur 3 to 4 bps from the 5′ edge of the protospacer. Additional on-target and off-target indel histogram plots are shown in Supplementary Fig. 4. (b)-(f) Scatterplots of indel frequencies (x-axis) and GUIDE-Seq sequencing read counts (y-axis) for cleavage sites identified by GUIDE-Seq for RGNs targeted to: VEGFA site 1, VEGFA site 2, VEGFA site 3, EMX1, and FANCF. (g) Fraction of potential RGN off-target sites bearing a certain number of mismatches that are cleaved (as detected by GUIDE-Seq). (h) Plots of GUIDE-Seq read counts (log-scale) for RGN off-target cleavage sites bearing a certain number of mismatches (i) Effects of mismatch position within the protospacer on GUIDE-Seq read counts for RGN off-target sites. Bases are numbered 1 to 20 with 20 being the base adjacent to the PAM. (j) Effects of wobble transition, non-wobble transition, and transversion mismatches estimated by linear regression analysis. (k) Fraction of GUIDE-Seq read count variance explained by individual univariate analyses for the effect of mismatch number, mismatch type, mismatch position, PAM density, expression level, and genomic position (intergenic/exon/intron).
Figure 4. Comparisons of GUIDE-Seq with computational prediction or ChIP-Seq methods for identifying RGN off-target sites
(a) Venn diagrams illustrating overlap between cleavage sites predicted by the MIT CRISPR Design Tool and GUIDE-Seq for nine RGNs. (b) Venn diagrams illustrating overlap between off-target sites predicted by the E-CRISP computational prediction program and GUIDE-Seq for nine RGNs. (c) Histogram showing the numbers of bona fide RGN off-target sites identified by GUIDE-Seq that are predicted, not predicted, and not considered by the MIT CRISPR Design Tool. Sites predicted by the MIT CRISPR Design Tool are divided into quintiles based on the score provided by the program. Each bar has the sites sub-classified based on the number of mismatches relative to the on-target site. Bulge sites are those that have a skipped base position at the gRNA-protospacer DNA interface. (d) Histogram showing the numbers of bona fide RGN off-target sites identified by GUIDE-Seq that are predicted, not predicted, and not considered by the E-CRISP computational prediction tool. Sites are subdivided as described in (c). (e) Venn diagrams illustrating overlap between dCas9 binding sites identified by ChIP-Seq and RGN off-target cleavage sites identified by GUIDE-Seq. (f) Histogram plots of RGN off-target sites identified by GUIDE-Seq and dCas9 binding sites identified by ChIP-Seq classified by the number of mismatches in the sequence relative to the intended on-target site. Kernel density estimation of GUIDE-Seq and ChIP-Seq mismatches is depicted. Dotted lines indicate the mean number of mismatches for each class of sites.
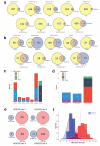
Figure 5. Large-scale structural alterations induced by RGNs
(a) Chromosome ideogram illustrating the locations of breakpoint hotspots in U2OS and HEK293 cells. Two hotspots overlap at the centromeric regions of chromosomes 1 and 10. (b) Schematic overview of AMP strategy for detecting translocations. Additional details in Online Methods. (c) Circos plots of structural variation induced by RGNs. Data for five RGNs and a control of cells are shown. Chromosomes are arranged in a circle with translocations shown as arcs between two chromosomal locations. Deletions or inversions greater than 1 kb in length are shown as straight lines. Sites that are not on-target, off-target, or breakpoint hotspots are classified as “other”. (d) Example of a translocation detected between the VEGFA site 1 on-target site on chromosome 6 and an off-target site on chromosome 17. All four possible reciprocal translocations were detected using AMP. (e) Examples of large deletion and inversion between two off-target sites in VEGFA site 2 detected by AMP. (f) Summary table of different RGN-induced and RGN-independent structural variations observed with five RGNs. Controls with Cas9 only, dsODN oligo only, and cells only are also shown.
Figure 6. GUIDE-Seq profiles of RGNs directed by tru-gRNAs
(a) Numbers of previously known and novel off-target cleavage sites identified for RGNs directed to the to VEGFA site 1, VEGFA site 3, and EMX1 target sites by matched full-length gRNAs and truncated gRNAs. Note that the data for the RGNs directed by full-length gRNAs are the same as those presented in Fig. 1e and is shown again here for ease of comparison. (b)-(d) Chromosome ideograms showing on- and off-target sites for RGNs directed to the VEGFA site 1, VEGFA site 3, and EMX1 target sites by matched full-length gRNAs and truncated gRNAs. Note that the ideograms for the RGNs directed by full-length gRNAs are the same as those presented in Fig. 1h and Supplementary Fig. 3 and are shown again here for ease of comparison. (e) GUIDE-Seq-based identification of DSBs induced by RGNs directed by tru-gRNAs. Mapped reads for the on-target sites of the three RGNs directed by tru-gRNAs we assessed by GUIDE-Seq are shown. In all cases, the target site sequence is shown with the 20 bp protospacer sequence to the left and the PAM sequence to the right on the x-axis. As with RGNs directed by full-length gRNAs, note how the highest peak falls within 3 to 4 bps of the 5′-edge of the NGG PAM sequence, the expected position of an RGN cleavage event. (f)-(h) Sequences of off-target sites identified by GUIDE-Seq for RGNs directed by tru-gRNAs. For each RGN, the intended target sequence is shown in the top line with cleaved sites shown underneath and with mismatches to the on-target site shown and highlighted in color. GUIDE-Seq sequencing read counts are shown to the right of each site. The intended on-target site is marked with a green square, previously known off-target sites of RGNs directed by both a full-length gRNA and a tru-gRNA are marked with a red diamond, and previously known off-target sites found only with RGNs directed by a tru-gRNA are marked with a yellow diamond. Previously known off-target sites were those that were shown to have a mutagenesis frequency of 0.1% or higher in an earlier report. Data is shown for RGNs directed by tru-gRNAs to the (f) VEGFA site 1, (g) VEGFA site 3, and (h) EMX1 target sites.
Comment in
- Mapping the precision of genome editing.
Gabriel R, von Kalle C, Schmidt M. Gabriel R, et al. Nat Biotechnol. 2015 Feb;33(2):150-2. doi: 10.1038/nbt.3142. Nat Biotechnol. 2015. PMID: 25658281 No abstract available.
Similar articles
- Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases.
Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW. Frock RL, et al. Nat Biotechnol. 2015 Feb;33(2):179-86. doi: 10.1038/nbt.3101. Epub 2014 Dec 15. Nat Biotechnol. 2015. PMID: 25503383 Free PMC article. - Tag-seq: a convenient and scalable method for genome-wide specificity assessment of CRISPR/Cas nucleases.
Huang H, Hu Y, Huang G, Ma S, Feng J, Wang D, Lin Y, Zhou J, Rong Z. Huang H, et al. Commun Biol. 2021 Jul 2;4(1):830. doi: 10.1038/s42003-021-02351-3. Commun Biol. 2021. PMID: 34215845 Free PMC article. - Defining genome-wide CRISPR-Cas genome-editing nuclease activity with GUIDE-seq.
Malinin NL, Lee G, Lazzarotto CR, Li Y, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Iafrate AJ, Le LP, Aryee MJ, Joung JK, Tsai SQ. Malinin NL, et al. Nat Protoc. 2021 Dec;16(12):5592-5615. doi: 10.1038/s41596-021-00626-x. Epub 2021 Nov 12. Nat Protoc. 2021. PMID: 34773119 Free PMC article. Review. - Massively targeted evaluation of therapeutic CRISPR off-targets in cells.
Pan X, Qu K, Yuan H, Xiang X, Anthon C, Pashkova L, Liang X, Han P, Corsi GI, Xu F, Liu P, Zhong J, Zhou Y, Ma T, Jiang H, Liu J, Wang J, Jessen N, Bolund L, Yang H, Xu X, Church GM, Gorodkin J, Lin L, Luo Y. Pan X, et al. Nat Commun. 2022 Jul 13;13(1):4049. doi: 10.1038/s41467-022-31543-6. Nat Commun. 2022. PMID: 35831290 Free PMC article. - Evaluating and Enhancing Target Specificity of Gene-Editing Nucleases and Deaminases.
Kim D, Luk K, Wolfe SA, Kim JS. Kim D, et al. Annu Rev Biochem. 2019 Jun 20;88:191-220. doi: 10.1146/annurev-biochem-013118-111730. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883196 Review.
Cited by
- Cleavage of DNA Substrate Containing Nucleotide Mismatch in the Complementary Region to sgRNA by Cas9 Endonuclease: Thermodynamic and Structural Features.
Baranova SV, Zhdanova PV, Koveshnikova AD, Pestryakov PE, Vokhtantsev IP, Chernonosov AA, Koval VV. Baranova SV, et al. Int J Mol Sci. 2024 Oct 9;25(19):10862. doi: 10.3390/ijms251910862. Int J Mol Sci. 2024. PMID: 39409191 Free PMC article. - TCR-T cell therapy: current development approaches, preclinical evaluation, and perspectives on regulatory challenges.
Golikova EA, Alshevskaya AA, Alrhmoun S, Sivitskaya NA, Sennikov SV. Golikova EA, et al. J Transl Med. 2024 Oct 4;22(1):897. doi: 10.1186/s12967-024-05703-9. J Transl Med. 2024. PMID: 39367419 Free PMC article. Review. - Effective genome editing with an enhanced ISDra2 TnpB system and deep learning-predicted ωRNAs.
Marquart KF, Mathis N, Mollaysa A, Müller S, Kissling L, Rothgangl T, Schmidheini L, Kulcsár PI, Allam A, Kaufmann MM, Matsushita M, Haenggi T, Cathomen T, Kopf M, Krauthammer M, Schwank G. Marquart KF, et al. Nat Methods. 2024 Nov;21(11):2084-2093. doi: 10.1038/s41592-024-02418-z. Epub 2024 Sep 23. Nat Methods. 2024. PMID: 39313558 - The Evolution of Nucleic Acid-Based Diagnosis Methods from the (pre-)CRISPR to CRISPR era and the Associated Machine/Deep Learning Approaches in Relevant RNA Design.
Chakraborty SS, Ray Dutta J, Ganesan R, Minary P. Chakraborty SS, et al. Methods Mol Biol. 2025;2847:241-300. doi: 10.1007/978-1-0716-4079-1_17. Methods Mol Biol. 2025. PMID: 39312149
References
Publication types
MeSH terms
Substances
Grants and funding
- DP1 GM105378/GM/NIGMS NIH HHS/United States
- DP1 GM105378/DP/NCCDPHP CDC HHS/United States
- R01 GM088040/GM/NIGMS NIH HHS/United States
- F32 GM105189/GM/NIGMS NIH HHS/United States
- R01 AR063070/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous