Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle - PubMed (original) (raw)
. 2015 Jan 6;112(1):244-9.
doi: 10.1073/pnas.1419038112. Epub 2014 Dec 22.
Jeffrey S McLean 2, Anna Edlund 3, Shibu Yooseph 4, Adam P Hall 4, Su-Yang Liu 5, Pieter C Dorrestein 6, Eduardo Esquenazi 7, Ryan C Hunter 8, Genhong Cheng 5, Karen E Nelson 4, Renate Lux 1, Wenyuan Shi 9
Affiliations
- PMID: 25535390
- PMCID: PMC4291631
- DOI: 10.1073/pnas.1419038112
Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle
Xuesong He et al. Proc Natl Acad Sci U S A. 2015.
Abstract
The candidate phylum TM7 is globally distributed and often associated with human inflammatory mucosal diseases. Despite its prevalence, the TM7 phylum remains recalcitrant to cultivation, making it one of the most enigmatic phyla known. In this study, we cultivated a TM7 phylotype (TM7x) from the human oral cavity. This extremely small coccus (200-300 nm) has a distinctive lifestyle not previously observed in human-associated microbes. It is an obligate epibiont of an Actinomyces odontolyticus strain (XH001) yet also has a parasitic phase, thereby killing its host. This first completed genome (705 kb) for a human-associated TM7 phylotype revealed a complete lack of amino acid biosynthetic capacity. Comparative genomics analyses with uncultivated environmental TM7 assemblies show remarkable conserved gene synteny and only minimal gene loss/gain that may have occurred as TM7x adapted to conditions within the human host. Transcriptomic and metabolomic profiles provided the first indications, to our knowledge, that there is signaling interaction between TM7x and XH001. Furthermore, the induction of TNF-α production in macrophages by XH001 was repressed in the presence of TM7x, suggesting its potential immune suppression ability. Overall, our data provide intriguing insights into the uncultivability, pathogenicity, and unique lifestyle of this previously uncharacterized oral TM7 phylotype.
Keywords: TM7; epibiont; human-associated; interspecies interaction; oral microbiome.
Conflict of interest statement
Conflict of interest statement: W.S. is a part-time chief science officer of C3 Jian, Inc., which has licensed technologies from the University of California Regents that could be indirectly related to this research project.
Figures
Fig. 1.
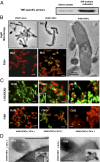
Cultivation and coisolation of TM7x with its host species Actinomyces spp. XH001. (A) PCR using a phylum-specific primer reveals the presence of TM7 within SHI medium-cultivated saliva samples. (B) Light microscopy and FISH images of XH100 monoculture (a and b) and XH100/TM7 coculture (c and d). (B, c) Cells of XH100 with several cells of TM7x attached to them. (B, d) Confocal laser scanning micrograph after hybridization with the cyanine5-labeled TM7567 (TM7x) and hexachloro-fluorescein–labeled universal eubacterial probe EUB338. XH001 appears red, whereas TM7 appears green. (B, e) TEM image of TM7x cell (indicated by arrows) attached to an XH001 cell. (Scale bars: 200 nm.) (C) Live/dead staining (a1, b1, and c1), and FISH (a2, b2, and c2) images. (C, a1 and a2) XH001/TM7x coculture 24 h after inoculation, with approximately two TM7x cells per XH001 cell. (C, b1 and b2) XH001/TM7x coculture 36 h after inoculation, with approximately six TM7x cells per XH001 cell. (C, b1, Inset) Live/dead staining of monoculture of XH001 36 h after inoculation. (C, c1 and c2) XH001/TM7x coculture 96 h after inoculation. For live/dead staining, live cells appear green and dead cells appear red; whereas for FISH, XH001 appears red and TM7x appears green. (D) TEM images showing the cell membrane of XH001 at/near the TM7x attachment site 24 h (a) and 36 h (b) after inoculation. (Insets) Original images from which D (a and b) are derived. (Scale bars: B, a_–_d and C, 1 μm; D, 200 nm.)
Fig. 2.
Host specificity of TM7x. Microscopic images of different Actinomyces species infected with isolated TM7x. Cultures were monitored under a microscope periodically up to 72 h, and random photographs were taken. Representative images are shown. (Scale bars: 1 μm.)
Fig. 3.
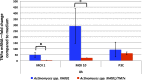
Induction of TNF-α production in macrophages by Actinomyces spp. XH001 monoculture and Actinomyces spp. XH001/TM7x coculture. Macrophages were treated with XH001 alone, XH001/TM7x coculture with a different multiplicity of infection (MOI), or Pam3CSK4 (P3C) as a positive control for 8 h. TNF-α mRNA was quantified using quantitative PCR. Fold induction was normalized to medium control. Each assay was performed in triplicate. Average values ± SD are shown. A Student t test (unpaired, two-tailed) was used for statistical analysis. An asterisk indicates a significant difference between the two values (P < 0.05).
Fig. 4.
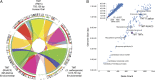
Comparative genomic analyses. (A) Genome synteny maintained between highly reduced genomes of human oral and environmentally derived TM7 phylum members. The RAAC3 and Saccharimonas aalborgensis genomes were assembled from environmental metagenomic reads into single contigs. The closed TM7x genome was assembled from an Actinomyces sp. (XH001) and TM7x symbiotic coculture. Related syntenic blocks between genomes are joined with colored ribbons revealing the larger number of syntenic regions and the maintained order of genes within each genome. From the inner ring to the outer ring, the colored blocks display the shared syntenic regions at increasing resolution. (B) Relationships between the microbial genome size of all finished genomes available in the Integrated Microbial Genomes Database (n = 2,086) and the number of predicted gene sequences. Smaller finished genomes are shown in the main panel. The three complete genomes representing members of the candidate phylum TM7 (TM7x, RAAC3, and S. aalborgensis) are marked with a color. (Inset) All microbial genomes that have less than 6 Mbp are shown, with the TM7x genome highlighted with a red marker.
Comment in
- Culture-based approaches to studying "microbial dark matter".
He X. He X. Proc Natl Acad Sci U S A. 2023 Jan 10;120(2):e2219691120. doi: 10.1073/pnas.2219691120. Epub 2023 Jan 3. Proc Natl Acad Sci U S A. 2023. PMID: 36595687 Free PMC article. No abstract available.
Similar articles
- Phenotypic and Physiological Characterization of the Epibiotic Interaction Between TM7x and Its Basibiont Actinomyces.
Bor B, Poweleit N, Bois JS, Cen L, Bedree JK, Zhou ZH, Gunsalus RP, Lux R, McLean JS, He X, Shi W. Bor B, et al. Microb Ecol. 2016 Jan;71(1):243-55. doi: 10.1007/s00248-015-0711-7. Epub 2015 Nov 23. Microb Ecol. 2016. PMID: 26597961 Free PMC article. - Quorum Sensing Modulates the Epibiotic-Parasitic Relationship Between Actinomyces odontolyticus and Its Saccharibacteria epibiont, a Nanosynbacter lyticus Strain, TM7x.
Bedree JK, Bor B, Cen L, Edlund A, Lux R, McLean JS, Shi W, He X. Bedree JK, et al. Front Microbiol. 2018 Sep 24;9:2049. doi: 10.3389/fmicb.2018.02049. eCollection 2018. Front Microbiol. 2018. PMID: 30319555 Free PMC article. - Transcriptome of Epibiont Saccharibacteria Nanosynbacter lyticus Strain TM7x During the Establishment of Symbiosis.
Hendrickson EL, Bor B, Kerns KA, Lamont EI, Chang Y, Liu J, Cen L, Schulte F, Hardt M, Shi W, He X, McLean JS. Hendrickson EL, et al. J Bacteriol. 2022 Sep 20;204(9):e0011222. doi: 10.1128/jb.00112-22. Epub 2022 Aug 17. J Bacteriol. 2022. PMID: 35975994 Free PMC article. - Saccharibacteria (TM7) in the Human Oral Microbiome.
Bor B, Bedree JK, Shi W, McLean JS, He X. Bor B, et al. J Dent Res. 2019 May;98(5):500-509. doi: 10.1177/0022034519831671. Epub 2019 Mar 20. J Dent Res. 2019. PMID: 30894042 Free PMC article. Review. - Genome evolution in bacterial endosymbionts of insects.
Wernegreen JJ. Wernegreen JJ. Nat Rev Genet. 2002 Nov;3(11):850-61. doi: 10.1038/nrg931. Nat Rev Genet. 2002. PMID: 12415315 Review.
Cited by
- Rich Repertoire of Quorum Sensing Protein Coding Sequences in CPR and DPANN Associated with Interspecies and Interkingdom Communication.
Bernard C, Lannes R, Li Y, Bapteste É, Lopez P. Bernard C, et al. mSystems. 2020 Oct 13;5(5):e00414-20. doi: 10.1128/mSystems.00414-20. mSystems. 2020. PMID: 33051376 Free PMC article. - RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria.
Wrighton KC, Castelle CJ, Varaljay VA, Satagopan S, Brown CT, Wilkins MJ, Thomas BC, Sharon I, Williams KH, Tabita FR, Banfield JF. Wrighton KC, et al. ISME J. 2016 Nov;10(11):2702-2714. doi: 10.1038/ismej.2016.53. Epub 2016 May 3. ISME J. 2016. PMID: 27137126 Free PMC article. - Microscopic and metatranscriptomic analyses revealed unique cross-domain parasitism between phylum Candidatus Patescibacteria/candidate phyla radiation and methanogenic archaea in anaerobic ecosystems.
Kuroda K, Nakajima M, Nakai R, Hirakata Y, Kagemasa S, Kubota K, Noguchi TQP, Yamamoto K, Satoh H, Nobu MK, Narihiro T. Kuroda K, et al. mBio. 2024 Mar 13;15(3):e0310223. doi: 10.1128/mbio.03102-23. Epub 2024 Feb 7. mBio. 2024. PMID: 38323857 Free PMC article. - Seminars in immunology special issue: Nutrition, microbiota and immunity The unexplored microbes in health and disease.
Plitt T, Faith JJ. Plitt T, et al. Semin Immunol. 2023 Mar;66:101735. doi: 10.1016/j.smim.2023.101735. Epub 2023 Feb 27. Semin Immunol. 2023. PMID: 36857892 Free PMC article. Review. - Into the darkness of the microbial dark matter in situ activities through expression profiles of Patescibacteria populations.
Vigneron A, Cruaud P, Guyoneaud R, Goñi-Urriza M. Vigneron A, et al. Front Microbiol. 2023 Jan 9;13:1073483. doi: 10.3389/fmicb.2022.1073483. eCollection 2022. Front Microbiol. 2023. PMID: 36699594 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 DE023810/DE/NIDCR NIH HHS/United States
- 1-R01-DE021108/DE/NIDCR NIH HHS/United States
- K99 DE024543/DE/NIDCR NIH HHS/United States
- R01 DE021108/DE/NIDCR NIH HHS/United States
- 1R01DE020102/DE/NIDCR NIH HHS/United States
- R01 DE020102/DE/NIDCR NIH HHS/United States
- 1R01DE023810-01/DE/NIDCR NIH HHS/United States
- K99DE024543/DE/NIDCR NIH HHS/United States
- R01 GM095373/GM/NIGMS NIH HHS/United States
- DE022734/DE/NIDCR NIH HHS/United States
- T90 DE022734/DE/NIDCR NIH HHS/United States
- 1R01GM095373/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases