Nodal signaling from the visceral endoderm is required to maintain Nodal gene expression in the epiblast and drive DVE/AVE migration - PubMed (original) (raw)
Nodal signaling from the visceral endoderm is required to maintain Nodal gene expression in the epiblast and drive DVE/AVE migration
Amit Kumar et al. Dev Biol. 2015.
Abstract
In the early mouse embryo, a specialized population of extraembryonic visceral endoderm (VE) cells called the distal VE (DVE) arises at the tip of the egg cylinder stage embryo and then asymmetrically migrates to the prospective anterior, recruiting additional distal cells. Upon migration these cells, called the anterior VE (AVE), establish the anterior posterior (AP) axis by restricting gastrulation-inducing signals to the opposite pole. The Nodal-signaling pathway has been shown to have a critical role in the generation and migration of the DVE/AVE. The Nodal gene is expressed in both the VE and in the pluripotent epiblast, which gives rise to the germ layers. Previous findings have provided conflicting evidence as to the relative importance of Nodal signaling from the epiblast vs. VE for AP patterning. Here we show that conditional mutagenesis of the Nodal gene specifically within the VE leads to reduced Nodal expression levels in the epiblast and incomplete or failed DVE/AVE migration. These results support a required role for VE Nodal to maintain normal levels of expression in the epiblast, and suggest signaling from both VE and epiblast is important for DVE/AVE migration.
Keywords: DVE/AVE; Epiblast; Nodal signaling; Ttr-Cre; Visceral endoderm.
Published by Elsevier Inc.
Figures
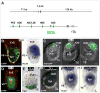
Fig. 1. Ttr-Cre activity coincides with first appearance of Nodal in VE
(A) Structure of the approximately 204 kB BAC transgene containing the 7.4 kb Nodal coding region, and 71 kb upstream and 126 kb downstream. Shown enlarged below is the area containing the three exons, with the coding region displayed in gray. The approximate positions of the Proximal Epiblast Enhancer (PEE) (Ben-Haim et al., 2006; Vincent et al., 2003), Node Specific Enhancer (NDE) (Adachi et al., 1999; Norris and Robertson, 1999), Asymmetric/Left Side-Specific Enhancer (AIE/LSE) (Saijoh et al., 2005; Vincent et al., 2004), Highly Bound Element (HBE) (Papanayotou et al., 2014) and Asymmetric Enhancer (ASE) (Norris et al., 2002; Norris and Robertson, 1999) are displayed as blue ellipses. The coding region within the first exon was replaced with EYFP followed by a stop codon. (B) Left panel, a projection of 6 consecutive confocal sections from an E6.0 transient transgenic embryo showing EYFP fluorescence in green and cell outlines marked in red with Alexa-Fluor 633 phalloidin. The scale bar represents 25 microns. Right panel, WMISH for Nodal expression of a similarly staged embryo. Expression in posterior epiblast (Epi) is seen in both embryos. Strong EYFP signal in VE is likely due to protein perdurance, as mRNA expression in VE is diminishing at this stage. (C) DIC images of E4.5 transgenic embryos superimposed with anti-GFP immunofluorescence signal (green) showing expression in single or multiple cells of the ICM. No signal is detected in the PrE and trophectoderm (TE). The scale bar represents 25 microns. (D) Confocal section of an E5.0 stage transgenic embryo showing EYFP fluorescence (green) in epiblast and distal VE. Cell outlines marked in red with Alexa-Fluor 633 phalloidin. The scale bar represents 25 microns. (E) Left panel, E5.0 embryo derived from Ttr-Cre by R26R cross. The majority of the VE shows X-gal stained cells reflecting Cre activity. Right panel, a similarly staged embryo from a Ttr-Cre by R26R-EFYP cross showing epifluorescence signal throughout the VE. (F) WMISH for Nodal expression in two E5.5 littermates derived from a Ttr-Cre, Nodal +/− by homozygous Nodal f l cross. The control embryo on the left is Nodal f l/− but Cre negative, and shows Nodal expression throughout the epiblast and VE. The conditional mutant embryo on the right is Nodal f l/− and Cre positive, and shows loss of Nodal expression from the VE.
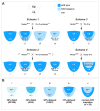
Fig. 2. Mating schemes and results for VE specific Nodal conditional mutants
(A) Mating schemes utilized for generating conditional mutants. The embryonic portion of E5.0 embryos is shown schematically with the epiblast and VE labeled in different colors depending on the possible genotype. Dark blue, wild type; light blue, heterozygous; pale, homozygous null. Scheme 1: Nodal f l/fl crossed to (X) Ttr-Cre, Nodal +/−. There are four potential genotypes of which (iv) represents the VE specific conditional mutant. The epiblast is heterozygous (fl/−) for Nodal. Scheme 2: Nodal f l/fl male X Ttr-Cre, Nodal +/fl female. There are four potential genotypes of which (iv) represents the VE specific conditional mutant. The epiblast is wild type (fl/fl) for Nodal. Scheme 3: Nodal f l/fl X Ttr-Cre. There are two potential genotypes of which (ii) represents the VE specific conditional heterozygote. The epiblast is wild type (+/fl) for Nodal. Scheme 4: Nodal f l/fl X Sox2-Cre. There are two potential genotypes of which (ii) represents the epiblast specific conditional heterozygote. The VE is wild type (+/fl) for Nodal. (B) The four different epiblast and VE genotypes resulting from conditional mutagenesis, as well as the Nodal heterozygote, are shown with their associated frequency of delayed or incomplete/failed AVE migration, color coded as for (A) above. Dark blue and light blue stippling represents reduced Nodal expression in the epiblast due to absence of Nodal signaling from VE.
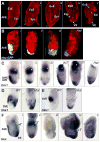
Fig. 3. AVE migration and other patterning defects in Scheme 1 conditional mutants
(A) Confocal sections of five E6.0 – E6.5 conditional mutants, stained with Alexa-Fluor 633 phalloidin to outline cells, showing no defects (i); a partially moved AVE (ii, asterisk); distal elongated VE cells (iii, asterisk); reduced epiblast growth (iv); overall reduced size and detached VE (v). The scale bar in i represents 25 microns; all embryos are at the same scale. (B) Confocal projections of an E6.5 wild type embryo (i), showing Hex-GFP marked AVE cells along the anterior; an E6.5 conditional mutant (ii), showing Hex-GFP marked cells partially moved; an E6.5 conditional mutant (iii), showing Hex-GFP marked cells all around the distal region; an E6.5 Nodal heterozygous embryo (Het), showing distal Hex-GFP marked cells. Cell outlines marked with Alexa-Fluor 633 phalloidin. The scale bar in i represents 25 microns; all embryos are at the same scale. (C) WMISH for Cerl mRNA expression in a late E5.5 wild type embryo (i), showing anterior location of DVE/AVE cells; a late E5.5 conditional mutant (ii), showing Cerl expressing cells only partially moved toward the anterior (bracket); a late E5.5 conditional mutant (iii), showing Cerl expressing cells all around the distal region (bracket); an E6.0 Nodal WT embryo showing Cerl expressing cells along the anterior; a heterozygous (Het) littermate showing distal Cerl expressing cells; another Het embryo showing Cerl expressing cells all around the distal region (bracket). White arrows in panels i – iii mark Esrrβ expression in the ExE. White arrowheads in the right panels mark Mash2 expression in the ExE. (D) Left, WMISH for Dkk1 mRNA expression in an early E6.0 wild type embryo. The distal region lacks expressing cells. Right, Dkk1 mRNA expression in an E6.0 conditional mutant embryo showing abnormal distal location of Dkk1 expressing cells. (E) Left panel, WMISH for Dkk1 mRNA expression in an E6.5 wild type embryo. Dkk1 expressing cells are found at the AVE. Right panel, Dkk1 mRNA expression in an E6.5 conditional mutant embryo showing abnormal distal location of Dkk1 expressing cells. (F) WMISH for Hex and T (Brachyury) mRNA expression in an E7.5 wild type embryo (i). Hex expressing cells (dark purple) are found in the AVE and definitive endoderm (DE). T expressing cells (light blue) mark mesoderm forming at the primitive streak. E7.5 conditional mutants are misshapen and can show distal Hex and proximal T expression (ii); lower Hex and T expression levels (iii); an expanded Hex expression domain with normal T levels (iv); reduced Hex and T expression domains (v).
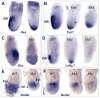
Fig. 4. AVE migration defects and reduced epiblast Nodal in Scheme 2 conditional mutants
(A) Two E6.5 littermates after WMISH for Hex mRNA. The wild type embryo (left) shows Hex in AVE cells while the conditional mutant shows distal Hex expression. (B) WMISH for Cerl mRNA in three E6.5 littermates. The wild type embryo (left) shows Cerl expression in the AVE while the conditional mutants (center, right) show distal expression. (C) WMISH for Goosecoid (Gsc) mRNA in two E6.5 littermates. The wild type embryo (left) shows Gsc in primitive streak cells at the posterior. The conditional mutant (right) shows only background levels and is smaller and misshapen. (D) Three E6.5 littermates following WMISH for Lefty1 mRNA expression. The wild type embryo (left) shows Lefty1 in the AVE while the conditional mutants show either distal (center) or no expression (right). (E) WMISH for Nodal mRNA in two early E5.5 littermates. The wild type embryo (left) shows Nodal throughout the epiblast and VE. The conditional mutant (right) lacks Nodal expression in the VE. (F) WMISH for Nodal mRNA expression in three E5.5 littermates. The wild type embryo (left) shows reduced Nodal in the distal epiblast and VE and strong proximal expression (white asterisk). The conditional mutants (right, center) completely lack Nodal expression in the VE and distal epiblast and show reduced proximal expression compared to wild type.
Similar articles
- Tissue-intrinsic beta-catenin signals antagonize Nodal-driven anterior visceral endoderm differentiation.
Schumacher S, Fernkorn M, Marten M, Chen R, Kim YS, Bedzhov I, Schröter C. Schumacher S, et al. Nat Commun. 2024 Jun 13;15(1):5055. doi: 10.1038/s41467-024-49380-0. Nat Commun. 2024. PMID: 38871742 Free PMC article. - Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning.
Mesnard D, Guzman-Ayala M, Constam DB. Mesnard D, et al. Development. 2006 Jul;133(13):2497-505. doi: 10.1242/dev.02413. Epub 2006 May 25. Development. 2006. PMID: 16728477 - The DVE changes distal epiblast fate from definitive endoderm to neurectoderm by antagonizing nodal signaling.
Miura S, Mishina Y. Miura S, et al. Dev Dyn. 2007 Jun;236(6):1602-10. doi: 10.1002/dvdy.21166. Dev Dyn. 2007. PMID: 17471538 - Dose-dependent Nodal/Smad signals pattern the early mouse embryo.
Robertson EJ. Robertson EJ. Semin Cell Dev Biol. 2014 Aug;32:73-9. doi: 10.1016/j.semcdb.2014.03.028. Epub 2014 Apr 1. Semin Cell Dev Biol. 2014. PMID: 24704361 Review. - Differential response of epiblast stem cells to Nodal and Activin signalling: a paradigm of early endoderm development in the embryo.
Kaufman-Francis K, Goh HN, Kojima Y, Studdert JB, Jones V, Power MD, Wilkie E, Teber E, Loebel DA, Tam PP. Kaufman-Francis K, et al. Philos Trans R Soc Lond B Biol Sci. 2014 Dec 5;369(1657):20130550. doi: 10.1098/rstb.2013.0550. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 25349457 Free PMC article. Review.
Cited by
- Mouse gastrulation: Coordination of tissue patterning, specification and diversification of cell fate.
Bardot ES, Hadjantonakis AK. Bardot ES, et al. Mech Dev. 2020 Sep;163:103617. doi: 10.1016/j.mod.2020.103617. Epub 2020 May 27. Mech Dev. 2020. PMID: 32473204 Free PMC article. Review. - JAM-A overexpression is related to disease progression in diffuse large B-cell lymphoma and downregulated by lenalidomide.
Xu PP, Sun YF, Fang Y, Song Q, Yan ZX, Chen Y, Jiang XF, Fei XC, Zhao Y, Leboeuf C, Li B, Wang CF, Janin A, Wang L, Zhao WL. Xu PP, et al. Sci Rep. 2017 Aug 7;7(1):7433. doi: 10.1038/s41598-017-07964-5. Sci Rep. 2017. PMID: 28785100 Free PMC article. - Engineering a computable epiblast for in silico modeling of developmental toxicity.
Barham K, Spencer R, Baker NC, Knudsen TB. Barham K, et al. Reprod Toxicol. 2024 Sep;128:108625. doi: 10.1016/j.reprotox.2024.108625. Epub 2024 Jun 8. Reprod Toxicol. 2024. PMID: 38857815 - Ino80 is essential for proximal-distal axis asymmetry in part by regulating Bmp4 expression.
Qiu Z, Elsayed Z, Peterkin V, Alkatib S, Bennett D, Landry JW. Qiu Z, et al. BMC Biol. 2016 Mar 14;14:18. doi: 10.1186/s12915-016-0238-5. BMC Biol. 2016. PMID: 26975355 Free PMC article. - Automated, High-Throughput Phenotypic Screening and Analysis Platform to Study Pre- and Post-Implantation Morphogenesis in Stem Cell-Derived Embryo-Like Structures.
Shankar V, van Blitterswijk C, Vrij E, Giselbrecht S. Shankar V, et al. Adv Sci (Weinh). 2024 Jan;11(4):e2304987. doi: 10.1002/advs.202304987. Epub 2023 Nov 22. Adv Sci (Weinh). 2024. PMID: 37991133 Free PMC article.
References
- Beck S, Le Good JA, Guzman M, Ben Haim N, Roy K, Beermann F, Constam DB. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat Cell Biol. 2002;4:981–985. - PubMed
- Ben-Haim N, Lu C, Guzman-Ayala M, Pescatore L, Mesnard D, Bischofberger M, Naef F, Robertson EJ, Constam DB. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell. 2006;11:313–323. - PubMed
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous