Mitochondrial protein quality control: the mechanisms guarding mitochondrial health - PubMed (original) (raw)
Review
Mitochondrial protein quality control: the mechanisms guarding mitochondrial health
Iryna Bohovych et al. Antioxid Redox Signal. 2015.
Abstract
Significance: Mitochondria are complex dynamic organelles pivotal for cellular physiology and human health. Failure to maintain mitochondrial health leads to numerous maladies that include late-onset neurodegenerative diseases and cardiovascular disorders. Furthermore, a decline in mitochondrial health is prevalent with aging. A set of evolutionary conserved mechanisms known as mitochondrial quality control (MQC) is involved in recognition and correction of the mitochondrial proteome.
Recent advances: Here, we review current knowledge and latest developments in MQC. We particularly focus on the proteolytic aspect of MQC and its impact on health and aging.
Critical issues: While our knowledge about MQC is steadily growing, critical gaps remain in the mechanistic understanding of how MQC modules sense damage and preserve mitochondrial welfare, particularly in higher organisms.
Future directions: Delineating how coordinated action of the MQC modules orchestrates physiological responses on both organellar and cellular levels will further elucidate the current picture of MQC's role and function in health, cellular stress, and degenerative diseases.
Figures
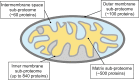
**FIG. 1.
Distribution of mitochondrial proteome throughout the organelle. The vast majority of mitochondrial proteins reside in the matrix and inner mitochondrial membrane (IM) subcompartments. The approximate numbers of polypeptides in each subproteome are calculated based on available data (82, 146, 156, 165, 185, 198). To see this illustration in color, the reader is referred to the web version of this article at
**FIG. 2.
Biochemical stresses that challenge normal mitochondrial function. Mitochondrial respiration is inherently linked to reactive oxygen species (ROS) production due to incomplete reduction of molecular oxygen by electron transport components of the oxidative phosphorylation system (OXPHOS system) (A). Stalling the high-energy electrons at respiratory complexes I and III leads to generation of superoxide anion which—either directly or via subsequent ROS radicals—can damage biological molecules like mtDNA and propel additional damage. The biogenesis of OXPHOS complexes requires tight coordination between synthesis and assembly of the mitochondrial- and nuclear-coded proteins (B). Polypeptides derived from the nuclear genome are translated on cytosolic ribosomes and imported in an unfolded state into the mitochondrion via presequence translocases of the outer (TOM) and inner (TIM) membranes. Imported polypeptides are inserted into the IM where they are joined with mitochondria-synthesized subunits. Mismatches in subunit stoichiometry can lead to accumulation of unfolded or unassembled proteins that can affect functional integrity of mitochondria. In addition, the electron transport chain units of OXPHOS contain redox-active cofactors poised for rapid electron exchange reactions (C). When improperly assembled, these prosthetic groups can act as pro-oxidants through their inherent ability to generate ROS via Fenton-like reactions (61, 161). To see this illustration in color, the reader is referred to the web version of this article at
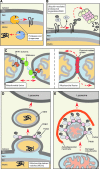
**FIG. 3.
Branches of the mitochondrial quality control (MQC) system. Multiple interdependent mechanisms exist at both molecular and organellar/cellular levels to sustain mitochondrial health. Conserved mitochondrial proteases and chaperones distributed across mitochondrial compartments represent one layer of MQC (A). Removal of the proteins localized to the outer mitochondrial membrane (OM) and potentially other mitochondrial subproteomes, termed mitochondria-associated degradation (MAD), is mediated by the cytosolic ubiquitin–proteasome system (UPS) and assisted by several E3 ubiquitin ligases (B). At the organellar level, MQC is provided through mitochondrial fusion (left panel C) and fission (right panel C) events, necessary for exchange and mixing of mitochondrial content and thus damage dilution, and segregation of damaged mitochondria from the network, respectively. Mitochondrial fusion is mediated by conserved GTPases in the OM (Mitofusins/Fzo1) and the IM (long and short isoforms of OPA1/Mgm1). Another OM-associated GTPase–Dynamin-related protein/Dnm1 is a key mediator of mitochondrial fission. Mitochondria-derived vesicles (MDVs), destined for lysosome, appear to represent yet another facet of organellar MQC (D). This mechanism allows selective removal of fragments of mitochondria without affecting the entire organelle. Reportedly, the MDVs contain oxidized cargo and lipids and their formation in mammalian cells depends on the function of PINK1 kinase and E3 ubiquitin ligase Parkin (see text for details). When mitochondrial damage overwhelms the aforementioned mechanisms, failing organelles are segregated and targeted to autophagosomes, and subsequently to lysosomes where their content is degraded. The PINK1-Parkin functional tandem and UPS play important roles in the initiation of this process known as mitophagy (E). To see this illustration in color, the reader is referred to the web version of this article at
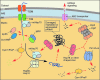
**FIG. 4.
PMQC in the matrix. Multiple proteases and molecular chaperones regulate the matrix subproteome. The regulation involves control of protein maturation and accumulation and degradation of poly- and oligopeptides. Proper maturation of the precursor proteins transported via the TIM23 translocase complex requires removal of mitochondrial targeting sequence by MPP processing metallopeptidase complex and, in certain cases, additional stabilizing processing by intermediate peptidases MIP/Oct1 and Icp55. Resulting free targeting peptides, as well as other small oligopeptides, are removed by mitochondrial presequence peptidase Cym1/PreP. Subsequent protein folding is facilitated by Hsp family chaperones. Stress-damaged, misfolded, and/or aggregated proteins are recognized and cleaved by AAA+ proteases Lon/Pim1 and ClpXP. Peptides produced by these proteolytic events are either subjected to additional processing by oligopeptidases or extruded through ATP-binding cassette (ABC)-type transporters into the cytosol where they activate mitochondrial unfolded protein response (UPRmt). To see this illustration in color, the reader is referred to the web version of this article at
**FIG. 5.
PMQC in the IM and intermembrane space (IMS). Complexity of mitochondrial IM anticipates vastly efficient systems to maintain protein homeostasis. These include two tightly coordinated proteases matrix-facing AAA metalloprotease (m-AAA) and intermembrane space-facing AAA metalloprotease (i-AAA), which along with other regulatory functions recognize excessive, misassembled, and damaged subunits of OXPHOS complexes associated with the IM. Another IM protease complex Oma1, with m-AAA-overlapping functions, is also proposed to play a major role in mitochondrial dynamics and homeostasis upon stress conditions. Rhomboid-like Pcp1/PARL protease is implicated in the intramembrane proteolysis of several IM proteins in yeast, whereas in mammalian cells, it also contributes to regulation of mitochondrial turnover. The IMS PMQC is less studied. In addition to the i-AAA, which exerts both proteolytic and chaperone functions toward IMS-localized proteins, the IMS subproteome appears to be regulated by oligopeptidase Prd1/Neurolysin and serine protease Ymn3/HtrA2. To see this illustration in color, the reader is referred to the web version of this article at
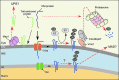
**FIG. 6.
PMQC in the OM. In addition to interception of mitochondria-destined proteins en route, the UPS provides an additional level of OM PMQC. It removes misfolded, damaged, or surplus proteins in the OM via the MAD process. MAD involves ubiquitylation by E3 ubiquitin ligases that tag proteins to be degraded and extraction of the peptides by the AAA+ protein VCP/Cdc48/p97 complex, which is, in turn, recruited to the OM through several mechanisms, including targeting by stress-responsive factor Vms1 or PINK1-Parkin functional tandem. Several reports (80, 123, 158) suggest that some IM proteins might also be subject of MAD. Another AAA+ protein Msp1/ATAD1 targets and removes tail-anchored proteins mislocalized to the OM. To see this illustration in color, the reader is referred to the web version of this article at
Similar articles
- Mitochondrial dysfunction in aging.
Guo Y, Guan T, Shafiq K, Yu Q, Jiao X, Na D, Li M, Zhang G, Kong J. Guo Y, et al. Ageing Res Rev. 2023 Jul;88:101955. doi: 10.1016/j.arr.2023.101955. Epub 2023 May 15. Ageing Res Rev. 2023. PMID: 37196864 Review. - Renal aging and mitochondrial quality control.
Guo X, Wang J, Wu Y, Zhu X, Xu L. Guo X, et al. Biogerontology. 2024 Jun;25(3):399-414. doi: 10.1007/s10522-023-10091-6. Epub 2024 Feb 13. Biogerontology. 2024. PMID: 38349436 Review. - The influence of aerobic exercise on mitochondrial quality control in skeletal muscle.
Philp AM, Saner NJ, Lazarou M, Ganley IG, Philp A. Philp AM, et al. J Physiol. 2021 Jul;599(14):3463-3476. doi: 10.1113/JP279411. Epub 2021 Jan 6. J Physiol. 2021. PMID: 33369731 - Fueling Inflamm-Aging through Mitochondrial Dysfunction: Mechanisms and Molecular Targets.
Picca A, Lezza AMS, Leeuwenburgh C, Pesce V, Calvani R, Landi F, Bernabei R, Marzetti E. Picca A, et al. Int J Mol Sci. 2017 Apr 28;18(5):933. doi: 10.3390/ijms18050933. Int J Mol Sci. 2017. PMID: 28452964 Free PMC article. Review. - Glucocorticoid impairs mitochondrial quality control in neurons.
Choi GE, Han HJ. Choi GE, et al. Neurobiol Dis. 2021 May;152:105301. doi: 10.1016/j.nbd.2021.105301. Epub 2021 Feb 17. Neurobiol Dis. 2021. PMID: 33609641 Review.
Cited by
- Translation stalling induced mitochondrial entrapment of ribosomal quality control related proteins offers cancer cell vulnerability.
Ojha R, Tantray I, Banerjee S, Rimal S, Thirunavukkarasu S, Srikrishna S, Chiu W, Mete U, Sharma A, Kakkar N, Lu B. Ojha R, et al. Res Sq [Preprint]. 2024 Sep 13:rs.3.rs-4899860. doi: 10.21203/rs.3.rs-4899860/v1. Res Sq. 2024. PMID: 39315278 Free PMC article. Preprint. - Targeting Mitochondrial COX-2 Enhances Chemosensitivity via Drp1-Dependent Remodeling of Mitochondrial Dynamics in Hepatocellular Carcinoma.
Che L, Wu JS, Du ZB, He YQ, Yang L, Lin JX, Lei Z, Chen XX, Guo DB, Li WG, Lin YC, Lin ZN. Che L, et al. Cancers (Basel). 2022 Feb 6;14(3):821. doi: 10.3390/cancers14030821. Cancers (Basel). 2022. PMID: 35159089 Free PMC article. - A mitochondrial unfolded protein response inhibitor suppresses prostate cancer growth in mice via HSP60.
Kumar R, Chaudhary AK, Woytash J, Inigo JR, Gokhale AA, Bshara W, Attwood K, Wang J, Spernyak JA, Rath E, Yadav N, Haller D, Goodrich DW, Tang DG, Chandra D. Kumar R, et al. J Clin Invest. 2022 Jul 1;132(13):e149906. doi: 10.1172/JCI149906. J Clin Invest. 2022. PMID: 35653190 Free PMC article. - Proteolytic regulation of mitochondrial dynamics.
Dietz JV, Bohovych I, Viana MP, Khalimonchuk O. Dietz JV, et al. Mitochondrion. 2019 Nov;49:289-304. doi: 10.1016/j.mito.2019.04.008. Epub 2019 Apr 25. Mitochondrion. 2019. PMID: 31029640 Free PMC article. Review. - Mechanisms of Mitochondrial Malfunction in Alzheimer's Disease: New Therapeutic Hope.
Nabi SU, Khan A, Siddiqui EM, Rehman MU, Alshahrani S, Arafah A, Mehan S, Alsaffar RM, Alexiou A, Shen B. Nabi SU, et al. Oxid Med Cell Longev. 2022 May 14;2022:4759963. doi: 10.1155/2022/4759963. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35607703 Free PMC article. Review.
References
- Abrahams BS, Mak GM, Berry ML, Palmquist DL, Saionz JR, Tay A, Tan YH, Brenner S, Simpson EM, and Venkatesh B. Novel vertebrate genes and putative regulatory elements identified at kidney disease and NR2E1/fierce loci. Genomics 80: 45–53, 2002 - PubMed
- Adam-Vizi V. and Chinopoulos C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol Sci 27: 639–645, 2006 - PubMed
- Alikhani N, Berglund AK, Engmann T, Spanning E, Vogtle FN, Pavlov P, Meisinger C, Langer T, and Glaser E. Targeting capacity and conservation of PreP homologues localization in mitochondria of different species. J Mol Biol 410: 400–410, 2011 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30GM103335/GM/NIGMS NIH HHS/United States
- 8 P20 GM103542-02/GM/NIGMS NIH HHS/United States
- R01 GM108975/GM/NIGMS NIH HHS/United States
- P20 GM103542/GM/NIGMS NIH HHS/United States
- 5P20RR024485-02/RR/NCRR NIH HHS/United States
- 1R01 GM108975/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical