Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment - PubMed (original) (raw)
Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment
Maorong Xie et al. J Virol. 2015 Mar.
Abstract
Recently, interferon-induced transmembrane proteins (IFITMs) have been identified to be key effector molecules in the host type I interferon defense system. The invasion of host cells by a large range of RNA viruses is inhibited by IFITMs during the entry step. However, the roles of IFITMs in DNA virus infections have not been studied in detail. In this study, we report that human cytomegalovirus (HCMV), a large human DNA virus, exploits IFITMs to facilitate the formation of the virion assembly compartment (vAC) during infection of human fibroblasts. We found that IFITMs were expressed constitutively in human embryonic lung fibroblasts (MRC5 cells). HCMV infection inhibited IFITM protein accumulation in the later stages of infection. Overexpression of an IFITM protein in MRC5 cells slightly enhanced HCMV production and knockdown of IFITMs by RNA interference reduced the virus titer by about 100-fold on day 8 postinfection, according to the findings of a virus yield assay at a low multiplicity of infection. Virus gene expression and DNA synthesis were not affected, but the typical round structure of the vAC was not formed after the suppression of IFITMs, thereby resulting in defective virion assembly and the production of less infectious virion particles. Interestingly, the replication of herpes simplex virus, a human herpesvirus that is closely related to HCMV, was not affected by the suppression of IFITMs in MRC5 cells. These results indicate that IFITMs are involved in a specific pathway required for HCMV replication.
Importance: HCMV is known to repurpose the interferon-stimulated genes (ISGs) viperin and tetherin to facilitate its replication. Our results expand the range of ISGs that can be exploited by HCMV for its replication. This is also the first report of a proviral function of IFITMs in DNA virus replication. In addition, whereas previous studies showed that IFITMs modulate virus entry, which is a very early stage in the virus life cycle, we identified a new function of IFITMs during the very late stage of virus replication, i.e., virion assembly. Virus entry and assembly both involve vesicle transport and membrane fusion; thus, a common biochemical activity of IFITMs is likely to be involved. Therefore, our findings may provide a new platform for dissecting the molecular mechanism of action of IFITMs during the blocking or enhancement of virus infection, which are under intense investigation in this field.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
FIG 1
HCMV infection suppresses IFITM gene expression. (A) MRC5 cells were mock infected or infected with HCMV at an MOI of 3, and the cell lysates were collected at the indicated times after infection. The protein levels of IFITM1 and IFITM2/3 were analyzed by immunoblotting. Commercially available antibodies recognize both IFITM2 and IFITM3 due to the high sequence similarities of the two proteins. Viral proteins IE1/2 were used to indicate HCMV infection, and tubulin was used as the loading control. (B) IFITM gene transcription was reduced by HCMV infection. MRC5 cells were mock infected or infected with HCMV at an MOI of 3. Total RNA was collected at the indicated times after infection. The mRNA levels of the IFITM genes were quantified by reverse transcription-qPCR. The relative gene expression level was normalized against that of GAPDH, and the normalized gene expression in mock-infected cells at 8 h was set equal to 1. (C) MRC5 cells were infected with HCMV (virus), UV-inactivated virus (virus + UV), or HCMV with ganciclovir (virus + GCV) at an MOI of 3 or mock infected (mock). Cell lysates were collected at 48 h after infection. (Left) The protein levels of IFITM1 and IFITM2/3 were analyzed by immunoblotting. Viral proteins IE1/2 and pp28 were used to indicate the effect of UV inactivation and GCV treatment on virus gene expression, and tubulin was used as the loading control. (Right) A longer exposure of anti-IFITM1.
FIG 2
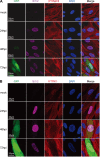
IFITM genes were suppressed by HCMV infection. (A) MRC5 cells were infected with HCMV (AD-GFP) at an MOI of 0.5 or mock infected. Cells were fixed at the indicated times after infection and stained with antibodies against the HCMV IE1/2 protein and the host IFITM2/3 protein. (B) An experiment similar to that described in the legend to panel A was performed, except that the cells were stained with IFITM1 and HCMV IE1/2 antibodies.
FIG 3
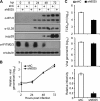
Overexpression of IFITMs in MRC5 cells fails to inhibit HCMV replication. (A) MRC5 cells were transduced with a lentiviral vector that expressed Flag-tagged IFITM1, IFITM2, or IFITM3 or a control (ctrl) vector, and the expression of IFITMs was analyzed at 48 h after transduction by immunoblotting using Flag tag-specific antibody. Tubulin was used as the loading control. (B) Growth analysis of HCMV infection in control or IFITM-expressing MRC5 cells at an MOI of 0.3. The cell-free virus in the supernatant was collected on the indicated days postinfection (dpi), and the titer was determined using a TCID50 assay. (C) Analysis of HCMV growth in control or IFITM-expressing MRC5 cells infected at an MOI of 3. (D) HCMV infection suppressed endogenous and overexpressed Flag-IFITM3. Control or Flag-IFITM3-expressing MRC5 cells were mock infected or infected with HCMV at an MOI of 3. Expression of IFITM3 was analyzed at 72 hpi by immunoblotting using IFITM3- or Flag tag-specific antibodies. Both Flag-tagged IFITM3 (arrowhead) and endogenous IFITM2/3 (arrow) were detected. (E) Overexpression of IFITMs did not affect HCMV protein expression. MRC5 cells overexpressing control or Flag-tagged IFITMs (left, IFITM1; middle, IFITM2; right, IFITM3) were infected with HCMV at an MOI of 3. Cell lysates were collected at the indicated times after infection. The accumulation of viral proteins with immediate early (IE1/2), early (UL38), and late (pp28) kinetics was examined by immunoblotting.
FIG 4
shRNA knockdown of IFITMs inhibits HCMV replication. (A) Knockdown efficiency of shRNAs targeting IFITM genes. Cells were transduced with lentivirus vectors expressing shC, sh8024, or sh8025. Cell lysates were collected at 48 h postransduction, and the protein levels of IFITM2/3 or IFITM1 were examined by immunoblotting. (B) Growth analysis of HCMV infection in cells expressing shC, sh8024, or sh8025. The cells were infected at an MOI of 0.3, the cell-free virus in the supernatant was collected at the indicated times after infection, and the titer was determined using a TCID50 assay. (C) Growth analysis of HCMV infection in cells expressing shC, sh8024, or sh8025 at an MOI of 3. The experimental procedures were similar to those described in the legend to panel B. (D) An independent experiment similar to that described in the legend to panel B, except the results were analyzed at more time points. (E) Downregulation of IFITM gene expression did not inhibit HSV replication. MRC5 cells were transduced with lentivirus overexpressing shC, sh8024, or sh8025 and then infected with HSV at an MOI of 0.01 or 0.001. The supernatant was collected at the indicated times postinfection, and titers were determined by a TCID50 assay. (F) Downregulation of IFITM gene expression did not induce elevated cell death. MRC5 cells were transduced with shC, sh8024, or sh8025 and then stained with propidium iodide (PI) and Hoechst at 3, 4, or 5 days after transduction. Hoechst stains the nucleus of all cells, and PI stains only dying cells with a permeable membrane. Cells treated with PBS for 5 h were included as a positive control for cell death.
FIG 5
Suppression of IFITM gene expression has a minimal effect on viral gene expression or DNA synthesis but significantly reduces the infectivity of the progeny virus particles. (A) Knockdown of IFITM gene expression by sh8025 had little effect on HCMV protein expression. MRC5 cells transduced with shC or sh8025 were infected with HCMV at an MOI of 3. Cell lysates were collected at the indicated times after infection. The accumulation of viral proteins with immediate early (IE1/2), early (UL38), and late (pp28) kinetics was examined by immunoblotting. (B) Knockdown of IFITM gene expression had a minimal effect on viral DNA synthesis. MRC5 cells transduced with shC or sh8025 were infected with HCMV at an MOI of 0.3. Intracellular DNA was extracted at the indicated times after infection. The accumulation of viral DNA was quantified by qPCR, and the amount was normalized against that of actin. The result for the DNA sample from shC-transduced cells collected at 2 h was set equal to 1. (C) Suppression of IFITM gene expression significantly reduced the infectivity of the progeny virus. shC- or sh8025-expressing cells were infected by HCMV at an MOI of 0.3. The cell-free virus in the culture medium was collected at 4 days p.i., concentrated, and purified by use of a sorbitol cushion. Each virus stock was divided into two to measure the infectious units by a TCID50 assay (top) and the genome copy number by qPCR (middle). (Bottom) The ratio of the TCID50 results relative to the DNA copy numbers was calculated for each sample. The ratio of the TCID50 results relative to the DNA copy number for the progeny virus produced in shC-expressing cells was set equal to 1.
FIG 6
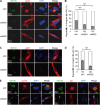
IFITMs are required for correct vAC formation. (A) MRC5 cells transduced with shC-, sh8024-, or sh8025-expressing lentiviruses were infected with HCMV (AD/Cre) at an MOI of 0.5. Cells were fixed at 96 h postinfection and stained with antibodies against virus structural protein pp28 and the Golgi apparatus marker GM130, both of which are proteins known to be localized to the vAC. DAPI was used to stain the nuclei. (B) Cells from the assay whose results are presented in panel A were counted for the presence of regular vAC and irregular vAC. A typical or regular vAC is characterized by a well-formed circular Golgi apparatus ring which is usually partially surrounded by kidney-like nuclei and a round pp28-positive structure. Irregular vAC displays a malformation of the circular Golgi apparatus ring and a scattered pp28 distribution. Statistical significance was measured with SPSS (version 20) software (SPSS Inc.) using the chi-square test. n, number of infected cells counted; ***, P < 0.002. (C) MRC5 cells transduced with shC- or sh8025-expressing lentiviruses were infected with HCMV (AD/Cre) at an MOI of 0.5. Cells were fixed at 72 h postinfection and stained with antibodies against virus structural protein pp28 and the Golgi apparatus marker GM130. DAPI was used to stain the nuclei. (D) Cells from the assay whose results are presented in panel C were counted for the presence of regular vAC and irregular vAC. n, number of infected cells counted; ***, P < 0.002. (E) MRC5 cells transduced with shC- or sh8025-expressing lentiviruses were infected with HCMV (AD/Cre) at an MOI of 0.5. Cells were fixed at 72 h postinfection and stained with antibodies against virus protein pp150 (left) or gB (right) and the Golgi apparatus marker GM130. DAPI was used to stain the nuclei.
FIG 7
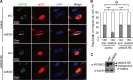
Overexpression of IFITM3 rescues vAC formation. (A) Control cells or shRNA-resistant IFITM3-expressing cells (sh8025-RE) were transduced with shC- or sh8025-expressing lentiviruses and then infected with HCMV (AD/Cre) at an MOI of 0.5. Cells were fixed at 72 h postinfection and stained with antibodies against virus structural protein pp28 and the Golgi apparatus marker GM130, both of which are proteins known to be localized to the vAC. DAPI was used to stain the nuclei. (B) Cells from the assay whose results are presented in panel A were counted for the presence of regular vAC and irregular vAC. Statistical significance was measured with SPSS (version 20) software (SPSS Inc.) using the chi-square test. n, number of infected cells counted; ns, no significant difference; ***, P < 0.002. (C) Western blotting of sh8025-resistant IFITM3 expression.
FIG 8
Electron microscopy analysis of the capsid assembly and DNA packaging in the presence or absence of IFITMs. (A) MRC5 cells were transduced with shC- or sh8025-expressing lentiviruses and then infected with HCMV (AD-GFP) at an MOI of 0.5, and cells were fixed at 96 hpi and prepared for electron microscopy. A capsids (white arrow), B capsids (white arrowhead), and C capsids (white arrowhead with a black border) are indicated. A capsids are devoid of viral DNA and a scaffold and presumably result from abortive virus DNA packaging events. B capsids contain a scaffold but lack viral DNA. C capsids contain the viral DNA and are devoid of a scaffold; thus, they likely represent the mature form of nucleocapsids. (B) Quantification of the different capsid types observed in the presence or absence of IFITMs (as average values per frame). The numbers of capsids counted were 187 for shC and 271 for sh8025.
FIG 9
Electron microscopy analysis of vAC formation and virion assembly reveals a defect in both processes after IFITM downregulation. shC- or sh8025-expressing MRC5 cells were infected with HCMV at an MOI of 0.5, and cells were fixed and stained at 96 hpi. Electron microscopic images of shC-expressing (A) and sh8025-expressing (B) cells infected with HCMV are shown to highlight the morphology of the nucleus (Nuc) and the vAC (circled). The white arrowhead with a black border points to the virion particles in the vAC. (C to E) Images of another infected control shRNA-expressing cell are shown at increasing magnifications to highlight vAC formation and virion assembly. The area in the square box in panel C is shown at a higher magnification in panel D, and the area in the square box in panel D is shown at a further increased magnification in panel E. Different forms of virion particles and dense bodies in panels D and E are indicated with arrows of different styles. (F to H) Images of another infected sh8025-expressing cell are shown at increasing magnifications. The areas in the square boxes labeled G and H in panel F are shown at higher magnifications in panels G and H, respectively. Arrow in panel G, the Golgi stacks; arrow in panel H, incompletely assembled virion particles without envelopment. (I) Quantification of HCMV secondary envelopment in the presence or absence of IFITMs. Enveloped virions are fully formed virions that contain DNA and envelope. Nonenveloped virions include naked particles and budding particles (virion particles attached to membranes). Statistical significance was measured using the chi-square test. n, number of virion particles counted; ***, P < 0.002.
Similar articles
- Identification of Host Factors Involved in Human Cytomegalovirus Replication, Assembly, and Egress Using a Two-Step Small Interfering RNA Screen.
McCormick D, Lin YT, Grey F. McCormick D, et al. mBio. 2018 Jun 26;9(3):e00716-18. doi: 10.1128/mBio.00716-18. mBio. 2018. PMID: 29946045 Free PMC article. - IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity.
Tartour K, Appourchaux R, Gaillard J, Nguyen XN, Durand S, Turpin J, Beaumont E, Roch E, Berger G, Mahieux R, Brand D, Roingeard P, Cimarelli A. Tartour K, et al. Retrovirology. 2014 Nov 25;11:103. doi: 10.1186/s12977-014-0103-y. Retrovirology. 2014. PMID: 25422070 Free PMC article. - Human Cytomegalovirus Hijacks WD Repeat Domain 11 for Virion Assembly Compartment Formation and Virion Morphogenesis.
Yang B, Yao Y, Cheng H, Wang XZ, Zhou YP, Huang SN, Ma X, Yang H, Wu J, Jiang X, Cheng S, Sun JY, Zeng WB, Chen J, Zhang FK, Shen HJ, Gu JY, McVoy MA, Britt WJ, Gong S, Luo MH. Yang B, et al. J Virol. 2022 Mar 9;96(5):e0182721. doi: 10.1128/JVI.01827-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35020472 Free PMC article. - [Interrelationship between human cytomegalovirus infection and chemokine].
Murayama T. Murayama T. Nihon Rinsho. 1998 Jan;56(1):69-74. Nihon Rinsho. 1998. PMID: 9465667 Review. Japanese. - Tegument proteins of human cytomegalovirus.
Kalejta RF. Kalejta RF. Microbiol Mol Biol Rev. 2008 Jun;72(2):249-65, table of contents. doi: 10.1128/MMBR.00040-07. Microbiol Mol Biol Rev. 2008. PMID: 18535146 Free PMC article. Review.
Cited by
- The Inhibition of HIV-1 Entry Imposed by Interferon Inducible Transmembrane Proteins Is Independent of Co-Receptor Usage.
Yu J, Liu SL. Yu J, et al. Viruses. 2018 Aug 7;10(8):413. doi: 10.3390/v10080413. Viruses. 2018. PMID: 30087232 Free PMC article. - Tuning the Orchestra: HCMV vs. Innate Immunity.
Dell'Oste V, Biolatti M, Galitska G, Griffante G, Gugliesi F, Pasquero S, Zingoni A, Cerboni C, De Andrea M. Dell'Oste V, et al. Front Microbiol. 2020 Apr 15;11:661. doi: 10.3389/fmicb.2020.00661. eCollection 2020. Front Microbiol. 2020. PMID: 32351486 Free PMC article. Review. - Localization of the WD repeat-containing protein 5 to the Virion Assembly Compartment Facilitates Human Cytomegalovirus Assembly.
Yang B, Yao Y, Wu H, Yang H, Ma XH, Li D, Wang XZ, Huang SN, Jiang X, Cheng S, Sun JY, Huang ZL, Zhao C, McVoy MA, Ahn JH, Zeng WB, Britt WJ, Gong S, Luo MH. Yang B, et al. J Virol. 2021 Mar 25;95(8):e02101-20. doi: 10.1128/JVI.02101-20. Epub 2021 Jan 27. J Virol. 2021. PMID: 33504601 Free PMC article. - Identification of Residues Controlling Restriction versus Enhancing Activities of IFITM Proteins on Entry of Human Coronaviruses.
Zhao X, Sehgal M, Hou Z, Cheng J, Shu S, Wu S, Guo F, Le Marchand SJ, Lin H, Chang J, Guo JT. Zhao X, et al. J Virol. 2018 Feb 26;92(6):e01535-17. doi: 10.1128/JVI.01535-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29263263 Free PMC article. - More than meets the I: the diverse antiviral and cellular functions of interferon-induced transmembrane proteins.
Shi G, Schwartz O, Compton AA. Shi G, et al. Retrovirology. 2017 Nov 21;14(1):53. doi: 10.1186/s12977-017-0377-y. Retrovirology. 2017. PMID: 29162141 Free PMC article. Review.
References
- Mocarski ES Jr, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegalovirus, p 1960–2014 InKnipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical