Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants - PubMed (original) (raw)
Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants
Ankita Bansal et al. Proc Natl Acad Sci U S A. 2015.
Abstract
Aging research has been very successful at identifying signaling pathways and evolutionarily conserved genes that extend lifespan with the assumption that an increase in lifespan will also increase healthspan. However, it is largely unknown whether we are extending the healthy time of life or simply prolonging a period of frailty with increased incidence of age-associated diseases. Here we use Caenorhabditis elegans, one of the premiere systems for lifespan studies, to determine whether lifespan and healthspan are intrinsically correlated. We conducted multiple cellular and organismal assays on wild type as well as four long-lived mutants (insulin/insulin-like growth factor-1, dietary restriction, protein translation, mitochondrial signaling) in a longitudinal manner to determine the health of the animals as they age. We find that some long-lived mutants performed better than wild type when measured chronologically (number of days). However, all long-lived mutants increased the proportion of time spent in a frail state. Together, these data suggest that lifespan can no longer be the sole parameter of interest and reveal the importance of evaluating multiple healthspan parameters for future studies on antiaging interventions.
Keywords: functional capacity; gerospan; healthspan; healthy aging; lifespan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
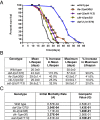
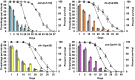
Fig. 1.
Survival analysis of the strains used in the study. (A) Survival curve of all strains used in this study: wild type (black), daf-2(e1370) (blue), eat-2(ad1113) (purple), ife-2(ok306) (orange), and clk-1(qm30) (yellow). (B) All strains have different mean and maximal lifespan and show differences compared with wild type. (C) Gompertz analysis based on the mortality rate of the mutant strains compared with wild type. A, initial mortality rate; G, Gompertz variable. The values were derived from the survival curves. A lower G value indicates a slower aging rate.
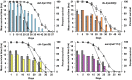
Fig. 2.
Resistance to heat stress decreases with age. Each graph represents a different long-lived mutant compared with wild type. The bar graph shows the average of the mean survival time under heat stress for a population of aging animals for two biological repeats, plotted on the primary (left) y axis. The dotted lines represent the percent survival of the individual strains (lifespan analysis), plotted on the secondary (right) y axis. Wild type is shown in each graph as gray bars and gray triangles. (Upper Left) daf-2(1370), blue bars and open circles. (Upper Right) ife-2(ok306), orange bars and open squares. (Lower Left) clk-1(qm30), yellow bars and open diamonds. (Lower Right) eat-2(ad1113), pink bars and open triangles. For all strains, resistance to heat was measured for different ages of each strain until 80% of the animals were dead and declines with age. At day 1, compared with wild type, clk-1, ife-2, and eat-2 mutants were sensitive to heat stress whereas daf-2 mutants were resistant. Compared with wild type, when analyzed chronologically, clk-1, ife-2, and daf-2 mutants show a similar rate of decline whereas eat-2 mutants show a faster rate of decline in oxidative stress resistance capacity. Compared with wild type, when analyzed physiologically, clk-1 and eat-2 mutants show a faster rate of decline whereas ife-2 and daf-2 mutants show a similar rate of decline in heat stress resistance capacity (see text for additional details). Error bars represent SE from two biological repeats.
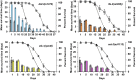
Fig. 3.
Resistance to oxidative stress decreases with age. Each graph represents a different long-lived mutant compared with wild type. The bar graph shows the average of the mean survival time under oxidative stress for a population of aging animals for two biological repeats, plotted on the primary (left) y axis. The dotted lines represent the percent survival of the individual strains (lifespan analysis), plotted on the secondary (right) y axis. Wild type is shown in each graph as gray bars and gray triangles. (Upper Left) daf-2(1370), blue bars and open circles. (Upper Right) ife-2(ok306), orange bars and open squares. (Lower Left) clk-1(qm30), yellow bars and open diamonds. (Lower Right) eat-2(ad1113), pink bars and open triangles. For all strains, oxidative stress resistance was measured for different ages of each strain until 80% of the animals were dead and declines with age. Compared with wild type chronologically, daf-2 mutants are more resistant to oxidative stress whereas ife-2 and clk-1 mutants are more sensitive to oxidative stress. Chronologically, compared with wild type, eat-2, ife-2, and daf-2 mutants show a similar rate of decline whereas clk-1 mutants have a slower rate of decline in oxidative stress resistance. Compared with wild type physiologically (age-matched population), ife-2 and clk-1 mutants show a similar rate of decline whereas daf-2 and eat-2 mutants show a faster rate of decline in oxidative stress resistance capacity (see text for additional details). Error bars represent SE from two biological repeats.
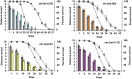
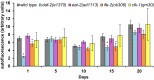
Fig. 4.
Movement capacity of worms: distance traveled on solid media. Each graph represents a different long-lived mutant compared with wild type. The bar graph shows the average distance traveled on solid media plates for each age plotted on the primary (left) y axis. An average of 15 worms was singled onto seeded NGM plates and allowed to move for 5 min. The distance traveled was measured (arbitrary units) and compared with wild type (gray filled bars). The dotted lines represent the percent survival of the individual strains (lifespan analysis), plotted on the secondary (right) y axis compared with wild type (gray triangles). Each graph represents a different long-lived mutant compared with wild type (gray bars and gray triangles). (Upper Left) daf-2(1370), blue bars and open circles. (Upper Right) ife-2(ok306), orange bars and open squares. (Lower Left) clk-1(qm30), yellow bars and open diamonds. (Lower Right) eat-2(ad1113), pink bars and open triangles. For all strains, the distance moved was measured for different ages of each strain until 80% of the animals were dead and declines with age. Compared with wild type, at day 1, eat-2 mutants cover significantly less distance whereas clk-1, daf-2, and ife-2 mutants move a similar distance. Chronologically, compared with wild type, daf-2 mutants’ movement capacity (distance) declined at the same rate, clk-1 and ife-2 mutants lost their movement capacity at a slower rate, and eat-2 mutants had a faster rate of decline. Physiologically (age-matched populations), compared with wild type, daf-2 and clk-1 mutants declined at the same rate, whereas ife-2 and eat-2 mutants lost their movement capacity faster (see text for additional details).
Fig. 5.
Movement capacity of aging worms: movement in liquid media. Each graph represents a different long-lived mutant compared with wild type. The bar graph indicates the average thrashings per minute for each age (15 animals total) plotted on the primary (left) y axis. The worms were transferred to M9 media and allowed to move in the liquid. The number of thrashes per minute was counted and then plotted. The dotted lines represent the percent survival of the individual strains (lifespan analysis), plotted on the secondary (right) y axis compared with wild type (gray triangles). Each graph represents a different long-lived mutant compared with wild type (gray bars and gray triangles). (Upper Left) daf-2(1370), blue bars and open circles. (Upper Right) ife-2(ok306), orange bars and open squares. (Lower Left) clk-1(qm30), yellow bars and open diamonds. (Lower Right) eat-2(ad1113), pink bars and open triangles. For all strains, movement was measured for different ages of each strain until 80% of the animals were dead and declines with age. Compared with wild type chronologically, daf-2 mutants thrash significantly less initially whereas clk-1 mutants move similar to wild type. The rate of decline in thrashing capacity is faster than wild type until young/midlife and then slows down at older age. When compared chronologically to wild type, the rate of decline is higher for eat-2 mutants and slower for ife-2 mutants. Compared with wild type physiologically, daf-2, clk-1, and eat-2 mutants show a faster rate of decline in movement capacity whereas for ife-2 mutants, movement capacity declines at the same rate.
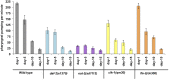
Fig. 6.
Pharyngeal pumping of aging worms. The bars represent the number of pharyngeal contractions per minute. Pumping rate declines with age for all of the strains until day 15. After day 15, all strains show negligible pumping. Error bars represent the SE for each strain (n = 10).
Fig. 7.
Changes in autofluorescence with age (lipofuscin). The bar graph indicates the changes in autofluorescence as the worms age. The amount of autofluorescence increased in all of the mutants. Compared with wild type, both daf-2 and ife-2 mutants show significantly lower accumulation at day 1 and day 15. After day 20, regardless of their mean/maximal lifespan, all of the strains show similar amounts of lipofuscin accumulation. *P < 0.05.
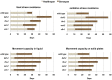
Fig. 8.
Comparison of chronological healthspan vs. gerospan ratio. Healthspan is defined as the period when the animal has greater than 50% of the maximal functional capacity of wild type. Gerospan is defined as the period when the animal has less than 50% of the maximal functional capacity of wild type. Comparisons were made by comparing the results for each day and are based on the maximal number of days each strain survives.
Fig. 9.
Comparison of physiological healthspan vs. gerospan ratio. Comparisons were made by normalizing the maximal lifespan to 100% for all strains.
Similar articles
- Movement decline across lifespan of Caenorhabditis elegans mutants in the insulin/insulin-like signaling pathway.
Newell Stamper BL, Cypser JR, Kechris K, Kitzenberg DA, Tedesco PM, Johnson TE. Newell Stamper BL, et al. Aging Cell. 2018 Feb;17(1):e12704. doi: 10.1111/acel.12704. Epub 2017 Dec 7. Aging Cell. 2018. PMID: 29214707 Free PMC article. - Royalactin extends lifespan of Caenorhabditis elegans through epidermal growth factor signaling.
Detienne G, De Haes W, Ernst UR, Schoofs L, Temmerman L. Detienne G, et al. Exp Gerontol. 2014 Dec;60:129-35. doi: 10.1016/j.exger.2014.09.021. Epub 2014 Oct 16. Exp Gerontol. 2014. PMID: 25456847 - Intrapopulation analysis of longitudinal lifespan in Caenorhabditis elegans identifies W09D10.4 as a novel AMPK-associated healthspan shortening factor.
Nakano Y, Moriuchi M, Fukushima Y, Hayashi K, Suico MA, Kai H, Koutaki G, Shuto T. Nakano Y, et al. J Pharmacol Sci. 2021 Mar;145(3):241-252. doi: 10.1016/j.jphs.2020.12.004. Epub 2020 Dec 29. J Pharmacol Sci. 2021. PMID: 33602504 - Two sides of lifespan regulating genes: pro-longevity or anti-longevity?
Honjoh S, Nishida E. Honjoh S, et al. J Biochem. 2011 Apr;149(4):381-8. doi: 10.1093/jb/mvr026. Epub 2011 Mar 3. J Biochem. 2011. PMID: 21372089 Review. - A pathway that links reproductive status to lifespan in Caenorhabditis elegans.
Kenyon C. Kenyon C. Ann N Y Acad Sci. 2010 Aug;1204:156-62. doi: 10.1111/j.1749-6632.2010.05640.x. Ann N Y Acad Sci. 2010. PMID: 20738286 Review.
Cited by
- Aging causes decreased resistance to multiple stresses and a failure to activate specific stress response pathways.
Dues DJ, Andrews EK, Schaar CE, Bergsma AL, Senchuk MM, Van Raamsdonk JM. Dues DJ, et al. Aging (Albany NY). 2016 Apr;8(4):777-95. doi: 10.18632/aging.100939. Aging (Albany NY). 2016. PMID: 27053445 Free PMC article. - Dietary protein restriction deciphers new relationships between lifespan, fecundity and activity levels in fruit flies Drosophila melanogaster.
Krittika S, Yadav P. Krittika S, et al. Sci Rep. 2020 Jun 22;10(1):10019. doi: 10.1038/s41598-020-66372-4. Sci Rep. 2020. PMID: 32572062 Free PMC article. - Age-associated vulval integrity is an important marker of nematode healthspan.
Leiser SF, Jafari G, Primitivo M, Sutphin GL, Dong J, Leonard A, Fletcher M, Kaeberlein M. Leiser SF, et al. Age (Dordr). 2016 Dec;38(5-6):419-431. doi: 10.1007/s11357-016-9936-8. Epub 2016 Aug 26. Age (Dordr). 2016. PMID: 27566309 Free PMC article. - Modeling neurodegeneration in Caenorhabditis elegans.
Caldwell KA, Willicott CW, Caldwell GA. Caldwell KA, et al. Dis Model Mech. 2020 Oct 26;13(10):dmm046110. doi: 10.1242/dmm.046110. Dis Model Mech. 2020. PMID: 33106318 Free PMC article. Review. - Bridging the Gap: A Geroscience Primer for Neuroscientists With Potential Collaborative Applications.
Hoffman JM, Hernandez CM, Hernandez AR, Bizon JL, Burke SN, Carter CS, Buford TW. Hoffman JM, et al. J Gerontol A Biol Sci Med Sci. 2022 Jan 7;77(1):e10-e18. doi: 10.1093/gerona/glab314. J Gerontol A Biol Sci Med Sci. 2022. PMID: 34653247 Free PMC article. Review.
References
- World Health Organization (2014) 10 Facts on Ageing and the Life Course. Available at www.who.int/features/factfiles/ageing/en. Accessed November 28, 2014.
- Meara E, White C, Cutler DM. Trends in medical spending by age, 1963–2000. Health Aff (Millwood) 2004;23(4):176–183. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources