Nur1 dephosphorylation confers positive feedback to mitotic exit phosphatase activation in budding yeast - PubMed (original) (raw)
Nur1 dephosphorylation confers positive feedback to mitotic exit phosphatase activation in budding yeast
Molly Godfrey et al. PLoS Genet. 2015.
Abstract
Substrate dephosphorylation by the cyclin-dependent kinase (Cdk)-opposing phosphatase, Cdc14, is vital for many events during budding yeast mitotic exit. Cdc14 is sequestered in the nucleolus through inhibitory binding to Net1, from which it is released in anaphase following Net1 phosphorylation. Initial Net1 phosphorylation depends on Cdk itself, in conjunction with proteins of the Cdc14 Early Anaphase Release (FEAR) network. Later on, the Mitotic Exit Network (MEN) signaling cascade maintains Cdc14 release. An important unresolved question is how Cdc14 activity can increase in early anaphase, while Cdk activity, that is required for Net1 phosphorylation, decreases and the MEN is not yet active. Here we show that the nuclear rim protein Nur1 interacts with Net1 and, in its Cdk phosphorylated form, inhibits Cdc14 release. Nur1 is dephosphorylated by Cdc14 in early anaphase, relieving the inhibition and promoting further Cdc14 release. Nur1 dephosphorylation thus describes a positive feedback loop in Cdc14 phosphatase activation during mitotic exit, required for faithful chromosome segregation and completion of the cell division cycle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
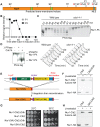
Figure 1. Nur1 dephosphorylation by Cdc14 in anaphase. A
. Schematic representation of Nur1. Putative (black) and confirmed (red) Cdk phosphorylation sites, as well as other predicted landmarks, are indicated. P1, SLpSPLRKpTPLSAR; P2, NDINSILRpSPK. B. Nur1 phosphopeptide abundance over the course of Cdc14-induced mitotic exit. Compare reference for details. C. Nur1 phosphorylation-dependent mobility shifts during synchronous cell cycle progression at 35.5°C, in wild-type and cdc14-1 cells, were analyzed using Phos-tag gels and Western blotting. Cell cycle progression was monitored by FACS analysis of DNA content. D. In vitro dephosphorylation of Nur1 by Cdc14. Immunopurified Nur1 was subjected to the indicated phosphatase or control treatments and analyzed by Phos-tag gel electrophoresis. E. Design scheme of the Nur1-Clb2 fusion strains. F. Western blot demonstrating replacement of endogenous Nur1 with Nur1-Clb2, or Nur1-Clb2ΔCdk, 4 hours after β-estradiol addition. G. Serial dilution assay to compare survival of Nur1-Clb2, Nur1-Clb2ΔCdk, Nur1(9A)-Clb2 and Nur1(9A)-Clb2ΔCdk-expressing cells, at 25°C and 36°C. H. Western blot demonstrating replacement of Nur1 or Nur1(9A) with Nur1-Clb2 and Nur1(9A)-Clb2, respectively, 4 hours after β-estradiol addition.
Figure 2. Nur1-Clb2 delays rDNA segregation.
A. rDNA segregation as a function of spindle length, compared between Nur1-Clb2 and Nur1-Clb2ΔCdk-expressing strains. At least 15, but typically more, cells were scored for each spindle length category. The mean and standard deviation from three independent experiments is shown. Wild type and Nur1(9A)-Cdk strains were also included in one repeat of the experiment. B. Representative images of rDNA segregation in a Nur1-Clb2ΔCdk cell, and of an anaphase bridge formed of undercondensed rDNA in a Nur1-Clb2 cell. The rDNA was visualized by the rDNA binding protein Net1, fused to YFP, the spindle was detected with an antibody against α-tubulin, DNA was stained with 4',6-diamidino-2-phenylindole (DAPI). C. ADE2 marker loss from the rDNA repeats is shown as a percentage of half red-sectored colonies after plating the indicated strains, with D. A representative field of colonies shown for each genotype. Around 500 colonies were scored for each strain. The mean and standard deviation from four independent experiments is shown.
Figure 3. Nur1-Clb2 causes a mitotic exit defect.
Nur1-Clb2 and Nur1-Clb2ΔCdk cells were synchronized in G1 by α-factor treatment and released to progress through the cell cycle at 36°C, before being rearrested in the following G1. At time points throughout the cell cycle we monitored, A. Cell cycle progression by FACS analysis of DNA content. B. Western blot analysis of the cell cycle markers securin, Clb2, Sic1 and Orc6, and C. The percentages of cells displaying metaphase (1–3 µm) or anaphase (>3 µm) spindles were scored. 100 cells were counted at each time point.
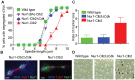
Figure 4. Nur1-Clb2 delays Cdc14 release.
A. Quantification of Cdc14 release, in experiments performed as in Fig. 3, relative to spindle length. At least 15, but typically more, cells were counted for each spindle length category. The mean and standard deviation from 3 independent experiments is shown. A logistic regression analysis showed that Cdc14 release timing in the Nur1-Clb2 strain was significantly delayed. B. Serial dilution assay demonstrating rescue of cell survival of Nur1-Clb2 cells by dominant active Cdc14TAB6-1. C. Western blot samples taken of the cells in B, before they were plated, shows that Cdc14TAB6-1 did not alter the Nur1-Clb2 phosphorylation status. D.-F. Active Cdc14 rescues mitotic progression of Nur1-Clb2 cells. As Fig. 3A-C, but strains carried the CDC14TAB6-1 allele.
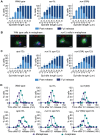
Figure 5. Phosphorylated Nur1 contributes to Cdc14 sequestration in early anaphase.
A. Quantification of Cdc14 release versus spindle length, as in Fig. 4A, in wild type, nur1Δ and nur1(9A) cells. Cdc14 release was subdivided into ‘Partial release’, when Cdc14 was detectably released into the nucleus but some nucleolar enrichment persisted and ‘Full release’, when no nucleolar Cdc14 enrichment remained detectable. The mean and standard deviation from three independent experiments is shown. B. Images of Cdc14 in wild type and nur1Δ cells. The metaphase state is confirmed by the presence of a short spindle, stained with an α-tubulin antibody. Cdc14-GFP was detected with an α-GFP antibody, DNA was counterstained with DAPI. C. As A, but cells carried compromised FEAR network activity due to the spo12Δ allele. D. Progression through mitosis of the strains above was measured by counting percentages of cells displaying metaphase (1–3 µm) or anaphase (>3 µm) spindles during synchronous cell cycle progression.
Figure 6. Nur1 acts in early anaphase and interacts with Net1.
A. Nur1 deletion does not reduce the requirement for the MEN. Strains of the indicated genotypes were streaked on YPD agar plates and grown at the temperatures shown. B. Nur1 interacts with Net1 throughout the cell cycle. Cell extracts were prepared from aliquots of a culture passing through a synchronous cell cycle at the indicated times. Co-immunoprecipitation of Nur1-Pk with Net1-myc was examined. A strain expressing Nur1-Pk but lacking the Net1-myc epitope served as a control. Cell cycle progression was monitored by FACS analysis of DNA content, the prevalent cell cycle stages are indicated.
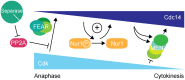
Figure 7. Nur1 establishes a positive feedback loop to promote Cdc14 release in early anaphase.
At the metaphase to anaphase transition, Cdc14 starts to be released following APC-mediated securin destruction, which liberates separase to downregulate PP2ACdc55. This allows Cdk to phosphorylate Net1 to initiate Cdc14 release. At the same time Cdk activity is in decline, thus removing the original impetus for Cdc14 release. To counteract this, Nur1 is dephosphorylated by early released Cdc14. This turns off Nur1's ability to inhibit Cdc14 and thus helps to sustain and augment Cdc14 release until the MEN pathway becomes active to maintain Cdc14 release while Cdk activity is further downregulated.
Similar articles
- Cdc14 Early Anaphase Release, FEAR, Is Limited to the Nucleus and Dispensable for Efficient Mitotic Exit.
Yellman CM, Roeder GS. Yellman CM, et al. PLoS One. 2015 Jun 19;10(6):e0128604. doi: 10.1371/journal.pone.0128604. eCollection 2015. PLoS One. 2015. PMID: 26090959 Free PMC article. - Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast.
Queralt E, Lehane C, Novak B, Uhlmann F. Queralt E, et al. Cell. 2006 May 19;125(4):719-32. doi: 10.1016/j.cell.2006.03.038. Cell. 2006. PMID: 16713564 - Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus.
Azzam R, Chen SL, Shou W, Mah AS, Alexandru G, Nasmyth K, Annan RS, Carr SA, Deshaies RJ. Azzam R, et al. Science. 2004 Jul 23;305(5683):516-9. doi: 10.1126/science.1099402. Science. 2004. PMID: 15273393 - At the interface between signaling and executing anaphase--Cdc14 and the FEAR network.
D'Amours D, Amon A. D'Amours D, et al. Genes Dev. 2004 Nov 1;18(21):2581-95. doi: 10.1101/gad.1247304. Genes Dev. 2004. PMID: 15520278 Review. - Regulation of CDC14: pathways and checkpoints of mitotic exit.
Bembenek J, Yu H. Bembenek J, et al. Front Biosci. 2003 Sep 1;8:d1275-87. doi: 10.2741/1128. Front Biosci. 2003. PMID: 12957817 Review.
Cited by
- Nucleolar release of rDNA repeats for repair involves SUMO-mediated untethering by the Cdc48/p97 segregase.
Capella M, Mandemaker IK, Martín Caballero L, den Brave F, Pfander B, Ladurner AG, Jentsch S, Braun S. Capella M, et al. Nat Commun. 2021 Aug 13;12(1):4918. doi: 10.1038/s41467-021-25205-2. Nat Commun. 2021. PMID: 34389719 Free PMC article. - Budding yeast relies on G1 cyclin specificity to couple cell cycle progression with morphogenetic development.
Pirincci Ercan D, Chrétien F, Chakravarty P, Flynn HR, Snijders AP, Uhlmann F. Pirincci Ercan D, et al. Sci Adv. 2021 Jun 4;7(23):eabg0007. doi: 10.1126/sciadv.abg0007. Print 2021 Jun. Sci Adv. 2021. PMID: 34088668 Free PMC article. - Analog-sensitive Cdk1 as a tool to study mitotic exit: protein phosphatase 1 is required downstream from Cdk1 inactivation in budding yeast.
Keaton JM, Workman BG, Xie L, Paulson JR. Keaton JM, et al. Chromosome Res. 2023 Sep 10;31(3):27. doi: 10.1007/s10577-023-09736-6. Chromosome Res. 2023. PMID: 37690059 - The Multiple Roles of the Cdc14 Phosphatase in Cell Cycle Control.
Manzano-López J, Monje-Casas F. Manzano-López J, et al. Int J Mol Sci. 2020 Jan 21;21(3):709. doi: 10.3390/ijms21030709. Int J Mol Sci. 2020. PMID: 31973188 Free PMC article. Review. - Exit from Mitosis in Budding Yeast: Protein Phosphatase 1 is Required Downstream from Cdk1 Inactivation.
Keaton JM, Workman BG, Xie L, Paulson JR. Keaton JM, et al. Res Sq [Preprint]. 2023 Apr 12:rs.3.rs-2787001. doi: 10.21203/rs.3.rs-2787001/v1. Res Sq. 2023. PMID: 37090579 Free PMC article. Updated. Preprint.
References
- Morgan D (2007) The cell cycle: Principles of control. London: New Science Press.
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, et al. (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 2: 709–718. - PubMed
- Bouchoux C, Uhlmann F (2011) A quantitative model for ordered Cdk substrate dephosphorylation during mitotic exit. Cell 147: 803–814. - PubMed
- Pereira G, Schiebel E (2003) Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302: 2120–2124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases