Identifying strains that contribute to complex diseases through the study of microbial inheritance - PubMed (original) (raw)
Identifying strains that contribute to complex diseases through the study of microbial inheritance
Jeremiah J Faith et al. Proc Natl Acad Sci U S A. 2015.
Abstract
It has been 35 y since Carl Woese reported in PNAS how sequencing ribosomal RNA genes could be used to distinguish the three domains of life on Earth. During the past decade, 16S rDNA sequencing has enabled the now frequent enumeration of bacterial communities that populate the bodies of humans representing different ages, cultural traditions, and health states. A challenge going forward is to quantify the contributions of community members to wellness, disease risk, and disease pathogenesis. Here, we explore a theoretical framework for studies of the inheritance of bacterial strains and discuss the advantages and disadvantages of various study designs for assessing the contribution of strains to complex diseases.
Keywords: disease; effector strains; health; microbial inheritance; strain-resolution human microbial ecology analyses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
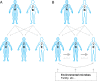
Fig. 1.
Human genetic inheritance and microbial inheritance. (A) In human genetic inheritance, information transmission is vertical, with 50% of each child’s genome content inherited from the father and 50% from the mother. Note that birth order is from left to right. (B) In microbial inheritance, transmission occurs largely in the first 3 y of life from proximal microbe-rich sources, notably family members, and in particular between near-birth siblings who provide a higher degree of access to their microbes. However, microbial inheritance is not limited to family members and can include other environmental sources, including family friends, nearby surfaces, and the local water supply.
Fig. 2.
Consequences of microbial inheritance. (A) As we age, the likelihood of transmitting and receiving microbial strains changes. The increased incidence of vomiting, diarrhea, and acute respiratory illnesses at early ages, combined with less established hygiene practices, likely enables young children to provide higher access to their microbial inhabitants. In addition, the less established community of microbes harbored in and on their bodies provides less resistance to invasion and establishment of new organisms. (B) The consequences of these age-associated changes in access and resistance are that the probability that a young child is colonized by a given microbial strain i [i.e., P(colonized i)] is higher when both parents harbor the strain than when only one parent does [i.e., P(access i) is higher because there are two low-access reservoirs of the strain rather than one], and higher if a near-birth sibling and a parent are colonized by the strain than if both parents are colonized [i.e., P(access i) is higher because there is one low-access reservoir and one high-access reservoir of the strain rather than two low-access reservoirs]. (C) In the context of multiple siblings, access is highest in near-birth siblings.
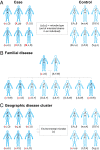
Fig. 3.
Study designs for identifying microbes that modulate complex disease risk. Microbial inheritance patterns provide an opportunity to identify etiologic agents of disease. By delineating the set of microbial inhabitants in each individual at the strain level, microbial inheritance patterns can be compared with disease incidence to identify strains whose presence/absence explains (correlates with) disease variation. As an illustrative example, every subject in this figure harbors a set of three microbial strains identified by their individual lower and uppercase letters (e.g., “a” is one strain and “A” is another). Healthy individuals are shown with a blue outline whereas affected individuals are shown in solid blue. Microbes that significantly increase disease risk are presented in red boldface (i.e., v, X, T, x, H, M). (A) A classic case-control design is difficult to power when searching for microbes that alter disease risk because, on average, unrelated individuals are expected to share no or very few strains. Sampling a broad enough population to have replicate observations of each strain would likely be prohibitively expensive. (B) Focusing on families increases the likelihood of identifying multiple individuals that share the same strain to enable identification of microbes enriched in either affected or unaffected individuals. Powering familial studies requires large families with multiple affected and unaffected individuals. (C) Geographic disease clusters provide a study design with high potential statistical power. Unrelated individuals are not expected to share microbial strains so identifying the same strain in multiple unrelated affected individuals would be highly significant and indicative of a shared environmental source of the identified strains.
Similar articles
- Computational integration of genomic traits into 16S rDNA microbiota sequencing studies.
Keller A, Horn H, Förster F, Schultz J. Keller A, et al. Gene. 2014 Oct 1;549(1):186-91. doi: 10.1016/j.gene.2014.07.066. Epub 2014 Jul 30. Gene. 2014. PMID: 25084126 - Assessing the Unseen Bacterial Diversity in Microbial Communities.
Caro-Quintero A, Ochman H. Caro-Quintero A, et al. Genome Biol Evol. 2015 Nov 27;7(12):3416-25. doi: 10.1093/gbe/evv234. Genome Biol Evol. 2015. PMID: 26615218 Free PMC article. - A quantitative SMRT cell sequencing method for ribosomal amplicons.
Jones BM, Kustka AB. Jones BM, et al. J Microbiol Methods. 2017 Apr;135:77-84. doi: 10.1016/j.mimet.2017.01.017. Epub 2017 Feb 1. J Microbiol Methods. 2017. PMID: 28159629 - Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories.
Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Woo PC, et al. Clin Microbiol Infect. 2008 Oct;14(10):908-34. doi: 10.1111/j.1469-0691.2008.02070.x. Clin Microbiol Infect. 2008. PMID: 18828852 Review. - A survey of the methods for the characterization of microbial consortia and communities.
Spiegelman D, Whissell G, Greer CW. Spiegelman D, et al. Can J Microbiol. 2005 May;51(5):355-86. doi: 10.1139/w05-003. Can J Microbiol. 2005. PMID: 16088332 Review.
Cited by
- The Host as the Driver of the Microbiota in the Gut and External Environment of Drosophila melanogaster.
Wong AC, Luo Y, Jing X, Franzenburg S, Bost A, Douglas AE. Wong AC, et al. Appl Environ Microbiol. 2015 Sep;81(18):6232-40. doi: 10.1128/AEM.01442-15. Epub 2015 Jul 6. Appl Environ Microbiol. 2015. PMID: 26150460 Free PMC article. - Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome.
Ussar S, Fujisaka S, Kahn CR. Ussar S, et al. Mol Metab. 2016 Jul 18;5(9):795-803. doi: 10.1016/j.molmet.2016.07.004. eCollection 2016 Sep. Mol Metab. 2016. PMID: 27617202 Free PMC article. Review. - Understanding the Role of the Gut Microbiome and Microbial Metabolites in Obesity and Obesity-Associated Metabolic Disorders: Current Evidence and Perspectives.
Vallianou N, Stratigou T, Christodoulatos GS, Dalamaga M. Vallianou N, et al. Curr Obes Rep. 2019 Sep;8(3):317-332. doi: 10.1007/s13679-019-00352-2. Curr Obes Rep. 2019. PMID: 31175629 Review. - HIV-associated changes in the enteric microbial community: potential role in loss of homeostasis and development of systemic inflammation.
Gootenberg DB, Paer JM, Luevano JM, Kwon DS. Gootenberg DB, et al. Curr Opin Infect Dis. 2017 Feb;30(1):31-43. doi: 10.1097/QCO.0000000000000341. Curr Opin Infect Dis. 2017. PMID: 27922852 Free PMC article. Review. - Diet-microbiome-disease: Investigating diet's influence on infectious disease resistance through alteration of the gut microbiome.
Harris EV, de Roode JC, Gerardo NM. Harris EV, et al. PLoS Pathog. 2019 Oct 31;15(10):e1007891. doi: 10.1371/journal.ppat.1007891. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31671152 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous