Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis - PubMed (original) (raw)
. 2015 Mar;35(6):988-1000.
doi: 10.1128/MCB.01242-14. Epub 2015 Jan 12.
Qingchao Tang 1, Dandan Qin 2, Lei Yu 1, Rui Huang 1, Guixiang Lv 3, Zhaoxia Zou 3, Xiao-chen Jiang 3, Chendan Zou 3, Wei Liu 3, Jing Luo 3, Zhixun Zhao 1, Shan Muhammad 1, Guiyu Wang 1, Ying-gang Chen 1, Xishan Wang 4
Affiliations
- PMID: 25582198
- PMCID: PMC4333091
- DOI: 10.1128/MCB.01242-14
Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis
Qian Zhang et al. Mol Cell Biol. 2015 Mar.
Retraction in
- Retraction for Zhang et al., "Role of MicroRNA 30a Targeting Insulin Receptor Substrate 2 in Colorectal Tumorigenesis".
Zhang Q, Tang Q, Qin D, Yu L, Huang R, Lv G, Zou Z, Jiang XC, Zou C, Liu W, Luo J, Zhao Z, Muhammad S, Wang G, Chen YG, Wang X. Zhang Q, et al. Mol Cell Biol. 2017 Jun 29;37(14):e00246-17. doi: 10.1128/MCB.00246-17. Print 2017 Jul 15. Mol Cell Biol. 2017. PMID: 28663275 Free PMC article. No abstract available.
Abstract
MicroRNAs (miRNAs) are dysregulated in many types of malignant diseases, including colorectal cancer. miRNA 30a (miR-30a) is a member of the miR-30 family and has been implicated in many types of cancers. In this study, we determined the expression of miR-30a in human colon cancer tissues and cell lines. miR-30a was found to be significantly downregulated in both the tissues and cell lines. Furthermore, overexpression of miR-30a inhibited, while silencing of miR-30a promoted, cell proliferation, migration, and invasion in vitro. Consistently, stable overexpression of miR-30a suppressed the growth of colon cancer cell xenografts in vivo. Moreover, bioinformatic algorithms and luciferase reporter assays revealed that insulin receptor substrate 2 (IRS2) is a direct target of miR-30a. Further functional studies suggested that repression of IRS2 by miR-30a partially mediated the tumor suppressor effect of miR-30a. In addition, miR-30a inhibited constitutive phosphorylation of Akt by targeting IRS2. Additionally, clinicopathological analysis indicated that miR-30a has an inverse correlation with the staging in patients with colon cancer. Taken together, our study provides the first evidence that miR-30a suppressed colon cancer cell growth through inhibition of IRS2. Thus, miR-30a might serve as a promising therapeutic strategy for colon cancer treatment.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
FIG 1
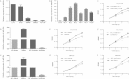
miR-30a suppressed cell growth ability in vitro. (A) Relative expression levels of miR-30a in five colon cancer cell lines were detected with the quantitative real-time PCR (qRT-PCR). Data represent the average of three independent experiments (error bars, standard errors). (B) The overexpression of miR-30a in HCT116 cell clones (no. 1 to no. 5) was determined by qRT-PCR. (C) Effects of stable overexpression of miR-30a on the proliferation of HCT116 cells were examined by MTT assay. Points are the average of three independent experiments; bars represent standard errors (*, P < 0.05; **, P < 0.01). (D) HCT116 cells were transiently transfected with an miR-30a mimic (or NC mimic) and inhibitor (or NC inhibitor). The expression of miR-30a was validated by qRT-PCR after 24 h. (E and F) Effects of transient overexpression and knockdown of miR-30a on the proliferation of HCT116 cells were examined by MTT assay. (G) Relative expression levels of miR-30a in SW620 cell lines transiently transfected with oligonucleotides were detected by qRT-PCR. (H and I) Effects of miR-30a on growth of SW620 cells were analyzed by MTT assay. (J and K) A colony formation assay was applied in mock, miR-30a pools, or HCT116 cells that were transiently transfected with miR-30a inhibitor or NC inhibitor (J), and the number of clones was quantitatively analyzed (K). The P values are relative to comparisons with NC mimic, mock, and NC inhibitor, from left to right. *, P < 0.05; **, P < 0.01. (L to O) Cell cycle distribution of HCT116 cells treated with miR-30a mimic (or NC mimic) and miR-30a inhibitor (or NC inhibitor) was assessed by flow cytometry 48 h posttransfection. Results are representative histogram of three independent experiments, plotting cell count versus DNA content. *, P < 0.05. OD490, optical density at 490 nm.
FIG 1
miR-30a suppressed cell growth ability in vitro. (A) Relative expression levels of miR-30a in five colon cancer cell lines were detected with the quantitative real-time PCR (qRT-PCR). Data represent the average of three independent experiments (error bars, standard errors). (B) The overexpression of miR-30a in HCT116 cell clones (no. 1 to no. 5) was determined by qRT-PCR. (C) Effects of stable overexpression of miR-30a on the proliferation of HCT116 cells were examined by MTT assay. Points are the average of three independent experiments; bars represent standard errors (*, P < 0.05; **, P < 0.01). (D) HCT116 cells were transiently transfected with an miR-30a mimic (or NC mimic) and inhibitor (or NC inhibitor). The expression of miR-30a was validated by qRT-PCR after 24 h. (E and F) Effects of transient overexpression and knockdown of miR-30a on the proliferation of HCT116 cells were examined by MTT assay. (G) Relative expression levels of miR-30a in SW620 cell lines transiently transfected with oligonucleotides were detected by qRT-PCR. (H and I) Effects of miR-30a on growth of SW620 cells were analyzed by MTT assay. (J and K) A colony formation assay was applied in mock, miR-30a pools, or HCT116 cells that were transiently transfected with miR-30a inhibitor or NC inhibitor (J), and the number of clones was quantitatively analyzed (K). The P values are relative to comparisons with NC mimic, mock, and NC inhibitor, from left to right. *, P < 0.05; **, P < 0.01. (L to O) Cell cycle distribution of HCT116 cells treated with miR-30a mimic (or NC mimic) and miR-30a inhibitor (or NC inhibitor) was assessed by flow cytometry 48 h posttransfection. Results are representative histogram of three independent experiments, plotting cell count versus DNA content. *, P < 0.05. OD490, optical density at 490 nm.
FIG 2
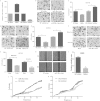
Overexpression of miR-30a inhibits cell migratory and invasive abilities. (A) HCT116 cells were transiently transfected with miR-30a mimic, NC mimic, inhibitor, NC inhibitor. The expression of miR-30a was determined by qRT-PCR after 24 h. (B and C) Effects of miR-30a on migration of HCT116 cells were analyzed by a transwell migration assay after 48 h. Representative photos (B) and quantitative analysis (C) are shown. (D and E) Effects of miR-30a on invasion of HCT116 cells were analyzed by a transwell invasion assay after 48 h. Representative photos (D) and quantitative analysis (E) are shown. (F and G) Effects of stable overexpression of miR-30a on migration and invasion were evaluated with transwell migration and invasion assays after 48 h. Representative photos (F) and quantitative analysis (G) are shown. (H) A wound-healing assay was performed to evaluate the effect of miR-30a on migration of SW620 cells. The artificial gap was through the central axis when cells reached a density of 80%. Photos of cells were taken at 0 and 48 h. (I) Relative migration length was from three randomly selected locations. (J and K) Real-time xCELLigence analysis of migration (represented by cell index) of HCT116 cells that were transiently transfected with miR-30a mimic, NC mimic, miR-30a inhibitor, or NC inhibitor. *, P < 0.05; **, P < 0.01.
FIG 3
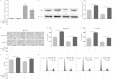
miR-30a directly targets the IRS2 3′ UTR. (A) The predicted targeting site with miR-30a of IRS2 3′ UTR. (B) miR-30a targeting sequences of IRS2 3′ UTR are evolutionarily conserved through six species (Hsa, human; Mml, rhesus monkey; Rno, rat; Bta, cow; Cfa, dog; Ptr, chimpanzee). The targeting sites are highlighted in bold. (C to E) qRT-PCR and Western blot analysis were applied to detect mRNA and protein expression of IRS2 in HCT116 cells that were stably transfected with pre-miR-30a (no. 1 and no. 3) or empty vector (mock). (F) IRS2 mRNA and miR-30a expression levels were determined in five colon cancer cell lines by qRT-PCR. (G) HEK293T cells were cotransfected with wild-type (WT 3′ UTR) or mutant (MUT 3′ UTR) reporters and the miR-30 family mimics or negative control (NC mimic). (H) HCT116 cells were cotransfected with wild-type or mutant reporters and the miR-30a mimic or negative control (NC mimic). In the experiments shown in both panels G and H, luciferase/Renilla activity was measured. *, P < 0.05; **, P < 0.01.
FIG 4
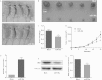
miR-30a suppressed colon cancer cell proliferation and migration through inhibition of IRS2 expression. (A to C) The expression levels of miR-30a and IRS2 were examined by qRT-PCR (A) and Western blot analysis (B and C), respectively, in HCT116 cells which were cotransfected with miR-30a mimic (or NC mimic) and IRS2 [or pcDNA3.1(+)]. β-Actin was used as the endogenous control. (D to G) The cells were subjected to transwell migration and MTT assays. Representative transwell pictures (D), quantitative analysis (E and F), and MTT results (G) are shown. (H and I) Cell cycle distribution of HCT116 cells was assessed by flow cytometry. Results are a representative histogram of three independent experiments, plotting cell count versus DNA content. *, P < 0.05; **, P < 0.01. (J and K) HCT116 cells were transfected with IRS2 siRNAs (IRS2-si-1 and IRS2-si-2) or an siRNA negative control (siNC). After 48 h, IRS2 expression was detected by Western blotting (J). Relative protein expression of IRS2, AKT, and p-AKT is shown, as indicated (K). Protein expression in the IRS2-si-1 and IRS2-si-2 groups was compared with that in the siNC group. **, P < 0.01. (L to N) After transfection with siRNA or siNC, HCT116 cells were subjected to transwell and MTT assays. Representative transwell pictures (L), quantitative analysis (M), and MTT results (N) are shown. (P and Q) Some certain proteins were detected by Western blotting in HCT116 cells that were transfected with miR-30a mimic (or NC mimic) and miR-30a inhibitor (or NC inhibitor). *, P < 0.05; **, P < 0.01.
FIG 4
miR-30a suppressed colon cancer cell proliferation and migration through inhibition of IRS2 expression. (A to C) The expression levels of miR-30a and IRS2 were examined by qRT-PCR (A) and Western blot analysis (B and C), respectively, in HCT116 cells which were cotransfected with miR-30a mimic (or NC mimic) and IRS2 [or pcDNA3.1(+)]. β-Actin was used as the endogenous control. (D to G) The cells were subjected to transwell migration and MTT assays. Representative transwell pictures (D), quantitative analysis (E and F), and MTT results (G) are shown. (H and I) Cell cycle distribution of HCT116 cells was assessed by flow cytometry. Results are a representative histogram of three independent experiments, plotting cell count versus DNA content. *, P < 0.05; **, P < 0.01. (J and K) HCT116 cells were transfected with IRS2 siRNAs (IRS2-si-1 and IRS2-si-2) or an siRNA negative control (siNC). After 48 h, IRS2 expression was detected by Western blotting (J). Relative protein expression of IRS2, AKT, and p-AKT is shown, as indicated (K). Protein expression in the IRS2-si-1 and IRS2-si-2 groups was compared with that in the siNC group. **, P < 0.01. (L to N) After transfection with siRNA or siNC, HCT116 cells were subjected to transwell and MTT assays. Representative transwell pictures (L), quantitative analysis (M), and MTT results (N) are shown. (P and Q) Some certain proteins were detected by Western blotting in HCT116 cells that were transfected with miR-30a mimic (or NC mimic) and miR-30a inhibitor (or NC inhibitor). *, P < 0.05; **, P < 0.01.
FIG 5
miR-30a inhibited tumorigenicity in a xenograft model. (A) A total of 5 × 106 HCT116 cells which were stably transfected with miR-30a or empty vector (mock) were subcutaneously injected into nude mice (n = 5). Mice were sacrificed 28 days after injection. (B) Tumors were harvested, and images of representative tumors are shown. (C) Tumors were weighed, and tumors in the miR-30a overexpression group weighed less than those of the mock group. (D) miR-30a overexpression resulted in inhibition of the growth rate. (E to G) The expression levels of miR-30a and IRS2 were detected by qRT-PCR (E) and Western blotting (F and G), respectively, in tumors. **, P < 0.01.
FIG 6
miR-30a is downregulated in colon cancer tissues and could function as a tumor suppressor in colon cancer. (A) miR-30a is downregulated in human colon cancer tissues. miR-30a expression was examined by qRT-PCR in 60 pairs of human colon cancer tissues. miR-30a expression was normalized to that of U6 in each sample. N, normal tissues; T, tumor tissues. (B) IRS2 mRNA and miR-30a levels were inversely correlated in colon cancer tissues as determined by qRT-PCR. β-Actin and U6 were used as the endogenous controls, respectively. (C to E) IRS2 was upregulated in colon cancer tissues compared with normal tissues, as indicated by immunohistochemistry (C) and Western blotting (D and E). (F) Kaplan-Meier curves illustrating correlation of miR-30a expression with overall survival (OS) (log rank test, P = 0.030). Patients with higher expression levels of miR-30a had better prognoses.
Similar articles
- MiR-30a-5p Suppresses Tumor Metastasis of Human Colorectal Cancer by Targeting ITGB3.
Wei W, Yang Y, Cai J, Cui K, Li RX, Wang H, Shang X, Wei D. Wei W, et al. Cell Physiol Biochem. 2016;39(3):1165-76. doi: 10.1159/000447823. Epub 2016 Aug 31. Cell Physiol Biochem. 2016. PMID: 27576787 - miR-20b-5p functions as tumor suppressor microRNA by targeting cyclinD1 in colon cancer.
Yang H, Lin J, Jiang J, Ji J, Wang C, Zhang J. Yang H, et al. Cell Cycle. 2020 Nov;19(21):2939-2954. doi: 10.1080/15384101.2020.1829824. Epub 2020 Oct 12. Cell Cycle. 2020. PMID: 33044899 Free PMC article. - MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA.
Wang Y, Tang Q, Li M, Jiang S, Wang X. Wang Y, et al. Biochem Biophys Res Commun. 2014 Feb 7;444(2):199-204. doi: 10.1016/j.bbrc.2014.01.028. Epub 2014 Jan 16. Biochem Biophys Res Commun. 2014. PMID: 24440701 - The Versatile Role of microRNA-30a in Human Cancer.
Yang X, Chen Y, Chen L. Yang X, et al. Cell Physiol Biochem. 2017;41(4):1616-1632. doi: 10.1159/000471111. Epub 2017 Mar 28. Cell Physiol Biochem. 2017. PMID: 28359057 Review. - miR-26 family and its target genes in tumorigenesis and development.
Li C, Li Y, Lu Y, Niu Z, Zhao H, Peng Y, Li M. Li C, et al. Crit Rev Oncol Hematol. 2021 Jan;157:103124. doi: 10.1016/j.critrevonc.2020.103124. Epub 2020 Oct 20. Crit Rev Oncol Hematol. 2021. PMID: 33254041 Review.
Cited by
- The IL-1β/AP-1/miR-30a/ADAMTS-5 axis regulates cartilage matrix degradation in human osteoarthritis.
Ji Q, Xu X, Zhang Q, Kang L, Xu Y, Zhang K, Li L, Liang Y, Hong T, Ye Q, Wang Y. Ji Q, et al. J Mol Med (Berl). 2016 Jul;94(7):771-85. doi: 10.1007/s00109-016-1418-z. Epub 2016 Apr 11. J Mol Med (Berl). 2016. PMID: 27067395 - DNA-Methylation-Caused Downregulation of miR-30 Contributes to the High Expression of XPO1 and the Aggressive Growth of Tumors in Pancreatic Ductal Adenocarcinoma.
Azmi AS, Li Y, Aboukameel A, Muqbil I, Philip PA, Mohammad RM. Azmi AS, et al. Cancers (Basel). 2019 Aug 2;11(8):1101. doi: 10.3390/cancers11081101. Cancers (Basel). 2019. PMID: 31382411 Free PMC article. - Overexpression of miR-30a in lung adenocarcinoma A549 cell line inhibits migration and invasion via targeting EYA2.
Yuan Y, Zheng S, Li Q, Xiang X, Gao T, Ran P, Sun L, Huang Q, Xie F, Du J, Xiao C. Yuan Y, et al. Acta Biochim Biophys Sin (Shanghai). 2016 Mar;48(3):220-8. doi: 10.1093/abbs/gmv139. Epub 2016 Feb 1. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 26837415 Free PMC article. - microRNA-141 inhibits thyroid cancer cell growth and metastasis by targeting insulin receptor substrate 2.
Dong S, Meng X, Xue S, Yan Z, Ren P, Liu J. Dong S, et al. Am J Transl Res. 2016 Mar 15;8(3):1471-81. eCollection 2016. Am J Transl Res. 2016. PMID: 27186273 Free PMC article. - MiR-30a: A Novel Biomarker and Potential Therapeutic Target for Cancer.
Jiang LH, Zhang HD, Tang JH. Jiang LH, et al. J Oncol. 2018 Aug 6;2018:5167829. doi: 10.1155/2018/5167829. eCollection 2018. J Oncol. 2018. PMID: 30158978 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical