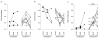Dysbiotic bacteria translocate in progressive SIV infection - PubMed (original) (raw)
Dysbiotic bacteria translocate in progressive SIV infection
Z Klase et al. Mucosal Immunol. 2015 Sep.
Abstract
Infection of gut-resident CD4(+) memory T cells during acute human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infection is associated with rapid loss of these cells and damage to the epithelial barrier. Damage to the epithelial barrier allows translocation of microbial products from the intestinal lumen into the body. Immune activation caused by these microbial products has been associated with disease progression. Although microbial translocation has been demonstrated in SIV-infected nonhuman primates, the identity of translocating bacteria has not been determined. In this study we examined the communities of bacteria both within the gastrointestinal (GI) tract and systemic tissues of both healthy and experimentally SIV-infected Asian macaques. Although there were only modest changes in the GI tract-associated microbiome resulting from infection, there is substantial dysbiosis after administration of antiretrovirals. Analysis of bacterial DNA isolated from tissues of infected animals revealed a preference for the phylum Proteobacteria, suggesting that they preferentially translocate. Consistent with this finding, we observed increased metabolic activity of Proteobacterial species within the colonic lumen of SIV-infected animals. Overall, these data provide insights into disease progression and suggest that therapies aimed at altering the composition and metabolic activity of the GI tract microbiome could benefit chronically HIV-infected individuals, particularly those on antiretroviral therapies.
Conflict of interest statement
Disclosure The authors declare that they have no conflict of interest.
Figures
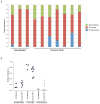
Figure 1. SIV and ARV treatment related changes in the GI tract microbiome
The GI tract microbiome of experimentally SIV-infected PTM was followed longitudinally by analyzing 16S rDNA content of stool. a) Stool DNA from day 0 and 21 post infection was amplified by PCR for 454 pyrosequencing analysis. Bar graphs indicate the relative makeup of the bacterial community in individual animals at each timepoint. b) The average bacterial community makeup of all 11 animals shown in panel A at day 0 and 21. Longitudinal qPCR analysis of the relative amount of c) Bacteroidetes, d) Firmictues, e) Proteobacteria or f) Lactobacillus present in the stool. Red dashed vertical line indicates the start of ARV therapy.
Figure 2. Changes in the metabolic activity of the microbiome
The metabolic activity of the GI tract microbiome of experimentally SIV-infected PTM was followed longitudinally by analyzing 16S RNA content of stool via RT-qPCR. a) The ratio of RNA/DNA for each of the three major phyla (Bacteroidetes, Firmicutes and Proteobacteria) for all animals prior to infection. Longitudinal RT-qPCR analysis of the relative metabolic activity of b) Bacteroidetes, c) Firmictues, d) Proteobacteria or e) Lactobacillus present in the stool. Red dashed vertical line indicates the start of ARV therapy.
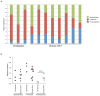
Figure 3. Analysis of the colon associated microbiome in SIV infection
DNA was extracted from the colon tissue of RM characterized as uninfected or SIV-infected. The relative abundance of Bacteroidetes, Firmicutes and Proteobacteria was determined by qPCR for 16S rDNA sequence. a) Bar graph showing the community structure of the colon microbiome for each of the animals. b) Dot plot showing the relative abundance of each phyla as in panel a.
Figure 4. Analysis of bacteria present in the mesenteric lymph node of SIV infected Rhesus macaques
DNA was extracted from the MLN of RM characterized as uninfected or SIV infected. The relative abundance of Bacteroidetes, Firmicutes and Proteobacteria was determined by qPCR for 16S rDNA sequence. a) Bar graph showing the community structure of bacteria from the MLN for each of the animals. b) Dot plot showing the relative abundance of each phyla as in panel a.
Figure 5. Abundance of tissue-associated Proteobacteria differs from the colon
The relative abundance of a) Bacteroidetes, b) Firmicutes and c) Proteobacteria in the colon, liver and MLN of uninfected or SIV-infected RM was determined by qPCR for Proteobacteria 16S rDNA.
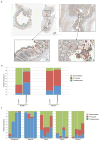
Figure 6. Specific families of proteobacteria translocate from the gut
DNA extracted from the colon, liver and MLN of uninfected and experimentally SIV-infected RM was amplified using bacterial 16S primers tagged for 454 pyro-sequencing analysis. a) Principal component analysis showing differences in the bacterial communities of the colon (blue) and liver (red) in SIV-infected (left panel) and uninfected (right panel) animals. b) Principal component analysis showing differences in the bacterial communities of the colon (blue) and MLN (green) in SIV-infected (left panel) and uninfected (right panel) animals. c) Metastats analysis was performed to determine which bacteria were present at significantly different abundance in either the liver or MLN compared to the colon of all RM or PTM used in the study. The graph shows bacterial families that are significantly under represented (blue) or over represented (red) in the tissues of each animal group.
Figure 7. Evidence for translocation of Proteobacteria from the colon
a) DNA was extracted from luminal swabs taken from the duodenum, jejunum, ileum, cecum and rectum from four uninfected RM. Relative abundance of Bacteroidetes, Firmicutes and Proteobacteria was determined by qPCR for 16S rDNA. b) To examine the makeup of bacteria present in the lamina propria of experimentally SIV-infected RM laser capture microdissection was performed on slide mounted colon tissue that had been stained with antibodies against bacterial lipopolysaccharide. Colored regions indicate samples taken from the lumen (blue), lamina propria (pink) and control (yellow). c) DNA was isolated from pooled samples from the laser capture and the relative abundance of Bacteroidetes, Firmicutes and Proteobacteria was determined for samples from the lumen or sites of translocation in the lamina propria by qPCR.
Figure 8. Correlation of proteobacteria translocation and activation of CD4+ T-cells
a) T cells from the MLN of RM were examined by flow cytometry to examine the percentage of CD4+ T-cells expressing the activation marker HLA-DR. Percentage of CD4+ T-cells expressing HLA-DR was plotted against the relative amount of Proteobacteria found in the MLN. b) Viral loads were determined at the time of necropsy and charted versus relative amount of Proteobacteria.
Similar articles
- IL-21 and probiotic therapy improve Th17 frequencies, microbial translocation, and microbiome in ARV-treated, SIV-infected macaques.
Ortiz AM, Klase ZA, DiNapoli SR, Vujkovic-Cvijin I, Carmack K, Perkins MR, Calantone N, Vinton CL, Riddick NE, Gallagher J, Klatt NR, McCune JM, Estes JD, Paiardini M, Brenchley JM. Ortiz AM, et al. Mucosal Immunol. 2016 Mar;9(2):458-67. doi: 10.1038/mi.2015.75. Epub 2015 Aug 19. Mucosal Immunol. 2016. PMID: 26286233 Free PMC article. - Microbial Dysbiosis During Simian Immunodeficiency Virus Infection is Partially Reverted with Combination Anti-retroviral Therapy.
Blum FC, Hardy BL, Bishop-Lilly KA, Frey KG, Hamilton T, Whitney JB, Lewis MG, Merrell DS, Mattapallil JJ. Blum FC, et al. Sci Rep. 2020 Apr 14;10(1):6387. doi: 10.1038/s41598-020-63196-0. Sci Rep. 2020. PMID: 32286417 Free PMC article. - Antiretroviral Therapy Administration in Healthy Rhesus Macaques Is Associated with Transient Shifts in Intestinal Bacterial Diversity and Modest Immunological Perturbations.
Ortiz AM, Flynn JK, DiNapoli SR, Sortino O, Vujkovic-Cvijin I, Belkaid Y, Sereti I, Brenchley JM. Ortiz AM, et al. J Virol. 2019 Aug 28;93(18):e00472-19. doi: 10.1128/JVI.00472-19. Print 2019 Sep 15. J Virol. 2019. PMID: 31270225 Free PMC article. - Microbial translocation in the pathogenesis of HIV infection and AIDS.
Marchetti G, Tincati C, Silvestri G. Marchetti G, et al. Clin Microbiol Rev. 2013 Jan;26(1):2-18. doi: 10.1128/CMR.00050-12. Clin Microbiol Rev. 2013. PMID: 23297256 Free PMC article. Review. - Mucosal immunity in human and simian immunodeficiency lentivirus infections.
Brenchley JM. Brenchley JM. Mucosal Immunol. 2013 Jul;6(4):657-65. doi: 10.1038/mi.2013.15. Epub 2013 Apr 3. Mucosal Immunol. 2013. PMID: 23549448 Free PMC article. Review.
Cited by
- The gut microbiome and HIV-1 pathogenesis: a two-way street.
Dillon SM, Frank DN, Wilson CC. Dillon SM, et al. AIDS. 2016 Nov 28;30(18):2737-2751. doi: 10.1097/QAD.0000000000001289. AIDS. 2016. PMID: 27755100 Free PMC article. Review. - Macrophages Are Phenotypically and Functionally Diverse across Tissues in Simian Immunodeficiency Virus-Infected and Uninfected Asian Macaques.
Ortiz AM, DiNapoli SR, Brenchley JM. Ortiz AM, et al. J Virol. 2015 Jun;89(11):5883-94. doi: 10.1128/JVI.00005-15. Epub 2015 Mar 18. J Virol. 2015. PMID: 25787286 Free PMC article. - Gut Microbiome, Short-Chain Fatty Acids, and Mucosa Injury in Young Adults with Human Immunodeficiency Virus Infection.
Qing Y, Xie H, Su C, Wang Y, Yu Q, Pang Q, Cui F. Qing Y, et al. Dig Dis Sci. 2019 Jul;64(7):1830-1843. doi: 10.1007/s10620-018-5428-2. Epub 2018 Dec 17. Dig Dis Sci. 2019. PMID: 30560340 - Brief Report: Inflammatory Colonic Innate Lymphoid Cells Are Increased During Untreated HIV-1 Infection and Associated With Markers of Gut Dysbiosis and Mucosal Immune Activation.
Dillon SM, Castleman MJ, Frank DN, Austin GL, Gianella S, Cogswell AC, Landay AL, Barker E, Wilson CC. Dillon SM, et al. J Acquir Immune Defic Syndr. 2017 Dec 1;76(4):431-437. doi: 10.1097/QAI.0000000000001523. J Acquir Immune Defic Syndr. 2017. PMID: 28825942 Free PMC article. - A Summary of the Fifth Annual Virology Education HIV Microbiome Workshop.
Sherrill-Mix S, Connors K, Aldrovandi GM, Brenchley JM, Boucher C, Bushman FD, Collman RG, Dandekar S, Klatt NR, Lagenaur LA, Paredes R, Tachedjian G, Turpin JA, Landay AL, Ghosh M. Sherrill-Mix S, et al. AIDS Res Hum Retroviruses. 2020 Nov;36(11):886-895. doi: 10.1089/AID.2020.0121. Epub 2020 Sep 7. AIDS Res Hum Retroviruses. 2020. PMID: 32777940 Free PMC article.
References
- Marchetti G, et al. Microbial translocation predicts disease progression of HIV-infected antiretroviral-naive patients with high CD4+ cell count. Aids. 2011 - PubMed
- Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials