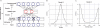Unsupervised feature construction and knowledge extraction from genome-wide assays of breast cancer with denoising autoencoders - PubMed (original) (raw)
Unsupervised feature construction and knowledge extraction from genome-wide assays of breast cancer with denoising autoencoders
Jie Tan et al. Pac Symp Biocomput. 2015.
Abstract
Big data bring new opportunities for methods that efficiently summarize and automatically extract knowledge from such compendia. While both supervised learning algorithms and unsupervised clustering algorithms have been successfully applied to biological data, they are either dependent on known biology or limited to discerning the most significant signals in the data. Here we present denoising autoencoders (DAs), which employ a data-defined learning objective independent of known biology, as a method to identify and extract complex patterns from genomic data. We evaluate the performance of DAs by applying them to a large collection of breast cancer gene expression data. Results show that DAs successfully construct features that contain both clinical and molecular information. There are features that represent tumor or normal samples, estrogen receptor (ER) status, and molecular subtypes. Features constructed by the autoencoder generalize to an independent dataset collected using a distinct experimental platform. By integrating data from ENCODE for feature interpretation, we discover a feature representing ER status through association with key transcription factors in breast cancer. We also identify a feature highly predictive of patient survival and it is enriched by FOXM1 signaling pathway. The features constructed by DAs are often bimodally distributed with one peak near zero and another near one, which facilitates discretization. In summary, we demonstrate that DAs effectively extract key biological principles from gene expression data and summarize them into constructed features with convenient properties.
Figures
Fig. 1
A) The network structure of denoising autoencoders. B) The distribution of one node's weight vector. C) The distribution of activity values for a node are bimodally distributed. Here we use Node5 as an example.
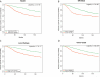
Fig. 2
Kaplan-Meier plots of disease-specific survival for Node5 (A), ER status (B), Luminal A subtype (C), and Tumor Grade (D) demonstrate that the constructed feature outperforms the other predictors.
Similar articles
- Association Analysis of Deep Genomic Features Extracted by Denoising Autoencoders in Breast Cancer.
Liu Q, Hu P. Liu Q, et al. Cancers (Basel). 2019 Apr 7;11(4):494. doi: 10.3390/cancers11040494. Cancers (Basel). 2019. PMID: 30959966 Free PMC article. - An unsupervised machine learning method for discovering patient clusters based on genetic signatures.
Lopez C, Tucker S, Salameh T, Tucker C. Lopez C, et al. J Biomed Inform. 2018 Sep;85:30-39. doi: 10.1016/j.jbi.2018.07.004. Epub 2018 Jul 29. J Biomed Inform. 2018. PMID: 30016722 Free PMC article. - Differential network analysis reveals the genome-wide landscape of estrogen receptor modulation in hormonal cancers.
Hsiao TH, Chiu YC, Hsu PY, Lu TP, Lai LC, Tsai MH, Huang TH, Chuang EY, Chen Y. Hsiao TH, et al. Sci Rep. 2016 Mar 14;6:23035. doi: 10.1038/srep23035. Sci Rep. 2016. PMID: 26972162 Free PMC article. - The Utility of Unsupervised Machine Learning in Anatomic Pathology.
McAlpine ED, Michelow P, Celik T. McAlpine ED, et al. Am J Clin Pathol. 2022 Jan 6;157(1):5-14. doi: 10.1093/ajcp/aqab085. Am J Clin Pathol. 2022. PMID: 34302331 Review. - Functional genomics and proteomics in the clinical neurosciences: data mining and bioinformatics.
Phan JH, Quo CF, Wang MD. Phan JH, et al. Prog Brain Res. 2006;158:83-108. doi: 10.1016/S0079-6123(06)58004-5. Prog Brain Res. 2006. PMID: 17027692 Review.
Cited by
- Wnt/_β_-Catenin, Carbohydrate Metabolism, and PI3K-Akt Signaling Pathway-Related Genes as Potential Cancer Predictors.
Chen P, Shi P, Du G, Zhang Z, Liu L. Chen P, et al. J Healthc Eng. 2019 Oct 20;2019:9724589. doi: 10.1155/2019/9724589. eCollection 2019. J Healthc Eng. 2019. PMID: 31781361 Free PMC article. - Exploring the druggable space around the Fanconi anemia pathway using machine learning and mechanistic models.
Esteban-Medina M, Peña-Chilet M, Loucera C, Dopazo J. Esteban-Medina M, et al. BMC Bioinformatics. 2019 Jul 2;20(1):370. doi: 10.1186/s12859-019-2969-0. BMC Bioinformatics. 2019. PMID: 31266445 Free PMC article. - Accurate Classification of Protein Subcellular Localization from High-Throughput Microscopy Images Using Deep Learning.
Pärnamaa T, Parts L. Pärnamaa T, et al. G3 (Bethesda). 2017 May 5;7(5):1385-1392. doi: 10.1534/g3.116.033654. G3 (Bethesda). 2017. PMID: 28391243 Free PMC article. - Developing and comparing deep learning and machine learning algorithms for osteoporosis risk prediction.
Qiu C, Su K, Luo Z, Tian Q, Zhao L, Wu L, Deng H, Shen H. Qiu C, et al. Front Artif Intell. 2024 Jun 11;7:1355287. doi: 10.3389/frai.2024.1355287. eCollection 2024. Front Artif Intell. 2024. PMID: 38919268 Free PMC article. - Deep Artificial Neural Networks and Neuromorphic Chips for Big Data Analysis: Pharmaceutical and Bioinformatics Applications.
Pastur-Romay LA, Cedrón F, Pazos A, Porto-Pazos AB. Pastur-Romay LA, et al. Int J Mol Sci. 2016 Aug 11;17(8):1313. doi: 10.3390/ijms17081313. Int J Mol Sci. 2016. PMID: 27529225 Free PMC article. Review.
References
- Vincent P, Larochelle H, Bengio Y, Manzagol P-A. Proceedings of the 25th International Conference on Machine Learning. ACM; New York, NY, USA: 2008. Extracting and composing robust features with denoising autoencoders.
- Bengio Y. Foundations and trends in Machine Learning. 2009;2:1.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous