scrm: efficiently simulating long sequences using the approximated coalescent with recombination - PubMed (original) (raw)
scrm: efficiently simulating long sequences using the approximated coalescent with recombination
Paul R Staab et al. Bioinformatics. 2015.
Abstract
Motivation: Coalescent-based simulation software for genomic sequences allows the efficient in silico generation of short- and medium-sized genetic sequences. However, the simulation of genome-size datasets as produced by next-generation sequencing is currently only possible using fairly crude approximations.
Results: We present the sequential coalescent with recombination model (SCRM), a new method that efficiently and accurately approximates the coalescent with recombination, closing the gap between current approximations and the exact model. We present an efficient implementation and show that it can simulate genomic-scale datasets with an essentially correct linkage structure.
© The Author 2015. Published by Oxford University Press.
Figures
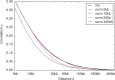
Fig. 1.
Approximation of genetic linkage. Shown is the correlation of ρ (_y_-axis) of the total local branch length at two sites δ base pairs apart (_x_-axis). The linkage in the CWR (ms, options
20 1 -r 4000 10000001 -T
) is indicated in black. Results for scrm using different exact window sizes (see legend) are indicated in colour
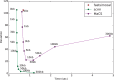
Fig. 2.
Efficiency for different approximations. Shown is the deviation (_y_-axis) against run-time (_x_-axis) for simulating 10 Mb with a recombination rate of 10−8 per base per generation. The deviation of the approximation from the correct values is measured as the square root of the area between the ρ−δ correlation curves for the approximate simulated data, and _ms_-generated data (see Fig. 1). For scrm and MaCS multiple approximation levels are drawn using different exact window sizes or history parameters. The recently published Cosi2 (Shlyakhter et al., 2014) does not output trees and could not be included in this figure; for a comparison of Cosi2 and scrm using different summary statistics see
Supplementary Figure S5
Similar articles
- Critical assessment of coalescent simulators in modeling recombination hotspots in genomic sequences.
Yang T, Deng HW, Niu T. Yang T, et al. BMC Bioinformatics. 2014 Jan 3;15:3. doi: 10.1186/1471-2105-15-3. BMC Bioinformatics. 2014. PMID: 24387001 Free PMC article. - The Bacterial Sequential Markov Coalescent.
De Maio N, Wilson DJ. De Maio N, et al. Genetics. 2017 May;206(1):333-343. doi: 10.1534/genetics.116.198796. Epub 2017 Mar 3. Genetics. 2017. PMID: 28258183 Free PMC article. - GENOMEPOP: a program to simulate genomes in populations.
Carvajal-Rodríguez A. Carvajal-Rodríguez A. BMC Bioinformatics. 2008 Apr 30;9:223. doi: 10.1186/1471-2105-9-223. BMC Bioinformatics. 2008. PMID: 18447924 Free PMC article. - Genome simulation approaches for synthesizing in silico datasets for human genomics.
Ritchie MD, Bush WS. Ritchie MD, et al. Adv Genet. 2010;72:1-24. doi: 10.1016/B978-0-12-380862-2.00001-1. Adv Genet. 2010. PMID: 21029846 Review. - Next-generation sequencing: big data meets high performance computing.
Schmidt B, Hildebrandt A. Schmidt B, et al. Drug Discov Today. 2017 Apr;22(4):712-717. doi: 10.1016/j.drudis.2017.01.014. Epub 2017 Feb 2. Drug Discov Today. 2017. PMID: 28163155 Review.
Cited by
- slendr: a framework for spatio-temporal population genomic simulations on geographic landscapes.
Petr M, Haller BC, Ralph PL, Racimo F. Petr M, et al. Peer Community J. 2023;3:e121. doi: 10.24072/pcjournal.354. Epub 2023 Dec 15. Peer Community J. 2023. PMID: 38984034 Free PMC article. - Accelerated Bayesian inference of population size history from recombining sequence data.
Terhorst J. Terhorst J. bioRxiv [Preprint]. 2024 Mar 27:2024.03.25.586640. doi: 10.1101/2024.03.25.586640. bioRxiv. 2024. PMID: 38585997 Free PMC article. Preprint. - Demes: a standard format for demographic models.
Gower G, Ragsdale AP, Bisschop G, Gutenkunst RN, Hartfield M, Noskova E, Schiffels S, Struck TJ, Kelleher J, Thornton KR. Gower G, et al. Genetics. 2022 Nov 1;222(3):iyac131. doi: 10.1093/genetics/iyac131. Genetics. 2022. PMID: 36173327 Free PMC article. - Mandrake: visualizing microbial population structure by embedding millions of genomes into a low-dimensional representation.
Lees JA, Tonkin-Hill G, Yang Z, Corander J. Lees JA, et al. Philos Trans R Soc Lond B Biol Sci. 2022 Oct 10;377(1861):20210237. doi: 10.1098/rstb.2021.0237. Epub 2022 Aug 22. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35989601 Free PMC article. - Approximate Bayesian Computation Untangles Signatures of Contemporary and Historical Hybridization between Two Endangered Species.
Dittberner H, Tellier A, de Meaux J. Dittberner H, et al. Mol Biol Evol. 2022 Feb 3;39(2):msac015. doi: 10.1093/molbev/msac015. Mol Biol Evol. 2022. PMID: 35084503 Free PMC article.
References
- Eriksson A., et al. . (2009) Sequential Markov coalescent algorithms for population models with demographic structure, Theor. Popul. Biol. , 76, 84–91. - PubMed
- Excoffier L., Foll M. (2011) fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios, Bioinformatics , 27, 1332–1334. - PubMed
- Hudson R.R. (2002) Generating samples under a Wright–Fisher neutral model, Bioinformatics, 18, 337–338. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous