Hybrid origin of European commercial pigs examined by an in-depth haplotype analysis on chromosome 1 - PubMed (original) (raw)
Hybrid origin of European commercial pigs examined by an in-depth haplotype analysis on chromosome 1
Mirte Bosse et al. Front Genet. 2015.
Abstract
Although all farm animals have an original source of domestication, a large variety of modern breeds exist that are phenotypically highly distinct from the ancestral wild population. This phenomenon can be the result of artificial selection or gene flow from other sources into the domesticated population. The Eurasian wild boar (Sus scrofa) has been domesticated at least twice in two geographically distinct regions during the Neolithic revolution when hunting shifted to farming. Prior to the establishment of the commercial European pig breeds we know today, some 200 years ago Chinese pigs were imported into Europe to improve local European pigs. Commercial European domesticated pigs are genetically more diverse than European wild boars, although historically the latter represents the source population for domestication. In this study we examine the cause of the higher diversity within the genomes of European commercial pigs compared to their wild ancestors by testing two different hypotheses. In the first hypothesis we consider that European commercial pigs are a mix of different European wild populations as a result of movement throughout Europe, hereby acquiring haplotypes from all over the European continent. As an alternative hypothesis, we examine whether the introgression of Asian haplotypes into European breeds during the Industrial Revolution caused the observed increase in diversity. By using re-sequence data for chromosome 1 of 136 pigs and wild boars, we show that an Asian introgression of about 20% into the genome of European commercial pigs explains the majority of the increase in genetic diversity. These findings confirm that the Asian hybridization, that was used to improve production traits of local breeds, left its signature in the genome of the commercial pigs we know today.
Keywords: Sus scrofa; domestication; genetic variation; haplotype homozygosity; hybridization; introgression.
Figures
Figure 1
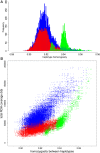
Neighbor-joining tree of all haplotypes of chromosome 1. Each individual has 2 haplotypes, one labeled after the name of the individual with the suffix “A” and the second haplotype contains the suffix “B.” Red line, Asian wild haplotype; orange line, Asian commercial or local haplotype; blue line, European commercial or local haplotype; green line, European wild haplotype. Locations of the markers on chromosome 1 are indicated by red bars. Alleles from the pig reference genome are included as two separate haplotypes without variation between them, and are highlighted in yellow.
Figure 2
Boxplots of homozygosity between two randomly paired haplotypes within groups. (1) Darkblue, two European local haplotypes; (2) blue, two European Iberian haplotypes; (3) lightblue, two European commercial haplotypes; (4) gray, two European wild haplotypes; (5) red, Asian commercial haplotypes; (6) brown, Asian local haplotypes; (7) orange, Asian wild haplotpyes (the highest dot indicates haplotype homozygosity within the Japanese wild boar).
Figure 3
Homozygosity between paired wild haplotypes. (A) Haplotype homozygosity between all possible pairs of European wild haplotypes. (B) Haplotype homozygosity between all possible pairs of 1 European wild and 1 Asian wild haplotye. (C) Haplotype homozygosity between all possible pairs of two Asian wild haplotypes.
Figure 4
Homozygosity between paired haplotypes in Europe. (A) Homozygosity between two European wild haplotypes is displayed in green. Homozygosity between two European commercial haplotypes is in red and the blue bars indicate homozygosity between one European wild and one European domesticated haplotype. (B) Homozygosity between haplotypes over the full chromosome on the x-axis is plotted against total ROH coverage between haplotypes on the y-axis for three combinations: two European commercial haplotypes (blue); two European wild haplotypes (green); one European wild and one European commercial haplotype (red).
Figure 5
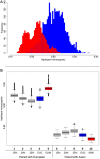
Haplotype homozygosity with Asian introgression. (A) Homozygosity between haplotypes when Asian commercial haplotypes are paired with European commercial (red) or European wild (blue). (B) Boxplots of haplotype homozygosity. Haplotypes are paired with European wild haplotypes (left) or Asian commercial haplotypes (right). Red boxes indicate haplotypes paired with European wild haplotypes. Blue boxes represent haplotypes that are paired with European commercial haplotypes. Gray boxes represent the distribution of homozygosity when the haplotype is paired with a chimeric haplotype that is a combination of a European wild haplotype and a Asian commercial haplotype (see also Box 1 in Supplementary material). (1) European wild paired with 15% Asian chimeric haplotype (2) European wild paired with 20% Asian chimeric haplotype (3) European wild paired with 25% Asian chimeric haplotype (4) European wild paired with European commercial (5) European wild paired with European wild (6) Asian commercial paired with 15% Asian chimeric haplotype (7) Asian commercial paired with 20% Asian chimeric haplotype (8) Asian commercial paired with 25% Asian chimeric haplotype (9) Asian commercial paired with European commercial (10) Asian commercial paired with European wild.
Similar articles
- Untangling the hybrid nature of modern pig genomes: a mosaic derived from biogeographically distinct and highly divergent Sus scrofa populations.
Bosse M, Megens HJ, Madsen O, Frantz LA, Paudel Y, Crooijmans RP, Groenen MA. Bosse M, et al. Mol Ecol. 2014 Aug;23(16):4089-102. doi: 10.1111/mec.12807. Epub 2014 Jun 16. Mol Ecol. 2014. PMID: 24863459 Free PMC article. - Genome-wide SNP data unveils the globalization of domesticated pigs.
Yang B, Cui L, Perez-Enciso M, Traspov A, Crooijmans RPMA, Zinovieva N, Schook LB, Archibald A, Gatphayak K, Knorr C, Triantafyllidis A, Alexandri P, Semiadi G, Hanotte O, Dias D, Dovč P, Uimari P, Iacolina L, Scandura M, Groenen MAM, Huang L, Megens HJ. Yang B, et al. Genet Sel Evol. 2017 Sep 21;49(1):71. doi: 10.1186/s12711-017-0345-y. Genet Sel Evol. 2017. PMID: 28934946 Free PMC article. - Molecular microevolution and epigenetic patterns of the long non-coding gene H19 show its potential function in pig domestication and breed divergence.
Li C, Wang X, Cai H, Fu Y, Luan Y, Wang W, Xiang H, Li C. Li C, et al. BMC Evol Biol. 2016 Apr 23;16:87. doi: 10.1186/s12862-016-0657-5. BMC Evol Biol. 2016. PMID: 27107967 Free PMC article. - RAPID COMMUNICATION: A haplotype information theory method reveals genes of evolutionary interest in European vs. Asian pigs.
Hudson NJ, Naval-Sánchez M, Porto-Neto L, Pérez-Enciso M, Reverter A. Hudson NJ, et al. J Anim Sci. 2018 Jul 28;96(8):3064-3069. doi: 10.1093/jas/sky225. J Anim Sci. 2018. PMID: 29873754 Free PMC article. - Artificial selection on introduced Asian haplotypes shaped the genetic architecture in European commercial pigs.
Bosse M, Lopes MS, Madsen O, Megens HJ, Crooijmans RP, Frantz LA, Harlizius B, Bastiaansen JW, Groenen MA. Bosse M, et al. Proc Biol Sci. 2015 Dec 22;282(1821):20152019. doi: 10.1098/rspb.2015.2019. Proc Biol Sci. 2015. PMID: 26702043 Free PMC article.
Cited by
- Selection of Optimal Ancestry Informative Markers for Classification and Ancestry Proportion Estimation in Pigs.
Liang Z, Bu L, Qin Y, Peng Y, Yang R, Zhao Y. Liang Z, et al. Front Genet. 2019 Mar 11;10:183. doi: 10.3389/fgene.2019.00183. eCollection 2019. Front Genet. 2019. PMID: 30915106 Free PMC article. - Adult porcine genome-wide DNA methylation patterns support pigs as a biomedical model.
Schachtschneider KM, Madsen O, Park C, Rund LA, Groenen MA, Schook LB. Schachtschneider KM, et al. BMC Genomics. 2015 Oct 5;16:743. doi: 10.1186/s12864-015-1938-x. BMC Genomics. 2015. PMID: 26438392 Free PMC article. - The Domestication of Wild Boar Could Result in a Relaxed Selection for Maintaining Olfactory Capacity.
Buglione M, Rivieccio E, Aceto S, Paturzo V, Biondi C, Fulgione D. Buglione M, et al. Life (Basel). 2024 Aug 22;14(8):1045. doi: 10.3390/life14081045. Life (Basel). 2024. PMID: 39202786 Free PMC article. - Genomic consequences of a century of inbreeding and isolation in the Danish wild boar population.
Yıldız B, Megens HJ, Hvilsom C, Bosse M. Yıldız B, et al. Evol Appl. 2022 May 17;15(6):954-966. doi: 10.1111/eva.13385. eCollection 2022 Jun. Evol Appl. 2022. PMID: 35782012 Free PMC article. - The pig pangenome provides insights into the roles of coding structural variations in genetic diversity and adaptation.
Li Z, Liu X, Wang C, Li Z, Jiang B, Zhang R, Tong L, Qu Y, He S, Chen H, Mao Y, Li Q, Pook T, Wu Y, Zan Y, Zhang H, Li L, Wen K, Chen Y. Li Z, et al. Genome Res. 2023 Oct;33(10):1833-1847. doi: 10.1101/gr.277638.122. Epub 2023 Nov 1. Genome Res. 2023. PMID: 37914227 Free PMC article.
References
- Alves P. C., Pinheiro I., Godinho R., Vicente J., Gortazar C., Scandura M. (2010). Genetic diversity of wild boar populations and domestic pig breeds (Sus scrofa) in South-western Europe. Biol. J. Linn. Soc. 101, 797–822 10.1111/j.1095-8312.2010.01530.x - DOI
- Bosse M., Megens H. J., Madsen O., Frantz L. A. F., Paudel Y., Crooijmans R. P. M. A., et al. . (2014b). Untangling the hybrid nature of modern pig genomes: a mosaic derived from biogeographically distinct and highly divergent Sus scrofa populations. Mol. Ecol. 23, 4089–4102. 10.1111/mec.12807 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials