Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin - PubMed (original) (raw)
Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin
Michal Schwartz et al. Nucleus. 2015.
Abstract
Nuclear pore complexes (NPCs) form the gateway to the nucleus, mediating virtually all nucleocytoplasmic trafficking. Assembly of a nuclear pore complex requires the organization of many soluble sub-complexes into a final massive structure embedded in the nuclear envelope. By use of a LacI/LacO reporter system, we were able to assess nucleoporin (Nup) interactions, show that they occur with a high level of specificity, and identify nucleoporins sufficient for initiation of the complex process of NPC assembly in vivo. Eleven nucleoporins from different sub-complexes were fused to LacI-CFP and transfected separately into a human cell line containing a stably integrated LacO DNA array. The LacI-Nup fusion proteins, which bound to the array, were examined for their ability to recruit endogenous nucleoporins to the intranuclear LacO site. Many could recruit nucleoporins of the same sub-complex and a number could also recruit other sub-complexes. Strikingly, Nup133 and Nup107 of the Nup107/160 subcomplex and Nup153 and Nup50 of the nuclear pore basket recruited a near full complement of nucleoporins to the LacO array. Furthermore, Nup133 and Nup153 efficiently targeted the LacO array to the nuclear periphery. Our data support a hierarchical, seeded assembly pathway and identify Nup133 and Nup153 as effective "seeds" for NPC assembly. In addition, we show that this system can be applied to functional studies of individual nucleoporin domains as well as to specific nucleoporin disease mutations. We find that the R391H cardiac arrhythmia/sudden death mutation of Nup155 prevents both its subcomplex assembly and nuclear rim targeting of the LacO array.
Keywords: ELYS; FG, phenylalanine-glycine; GFP, green fluorescent protein; CFP, cyan fluorescent protein; LacO, Lac operon; LacI, Lac repressor gene; LacI; LacO array; Nup133; Nup153; Nup155 R391H; Nup160/107 complex; cardiac arrhythmia; nuclear pore assembly; nuclear rim localization.
Figures
Figure 1.
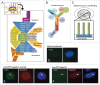
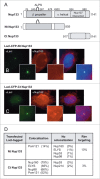
(A) A cut-away schematic representation of the massive nuclear pore complex, which has 8 radial spokes, is shown (inset). The subcomplexes that comprise one radial spoke and their predicted localization within that spoke are shown in the enlarged portion of (A). The nucleoporins used as LacI-CFP fusions in this study are highlighted in yellow. Each separate shape indicates a different subcomplex that must assemble with others to form a nuclear pore. The different colors are used to demarcate distinct regions of the formed nuclear pore, such as the large Y complex subunits (blue), ELYS (lavender), the central scaffold (green), the central transporter regions (gold), the cytoplasmic ring (purple), the cytoplasmic filaments (deep purple), and the nuclear basket (light brown). The three transmembrane Nups are shown in gray. (A few nucleoporins such as Aladin and hCG1 have been omitted for clarity). (B) Details of the Nup107–160 or “Y” complex, with the nucleoporins tested as LacI-CFP fusions underlined. (C) Diagram of the LacI-LacO system used in this study. A diagram of a U2OS 2–6–3 cell is shown, with the LacO array integrated into one location in the genome. The enlargement shows the expected binding of a fusion LacI-CFP-NupX (green) to each of the multiple copies of the LacO array (blue). (D–F) U2OS 2–6–3 cells were mock transfected (D), transfected with the negative control fusion LacI-CFP (E), or transfected with LacI-CFP-Nup fusion such as LacI-CFP Nup214 (F). To test for LacI-CFP-NupX binding to the LacO array, cells were visualized with anti-LacI to detect the fusion protein (left panel in each set), or a specific anti-nucleoporin antibody (middle panel in E and F). DNA was detected with DAPI stain. The right-hand panel in each set shows a merge of all 3 stains. The arrows point to the position of the LacO array.
Figure 2.
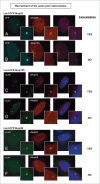
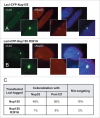
Nucleoporins anchored to the LacO array recruit other nucleoporins from the same subcomplex. Immunofluorescence microscopy of U2OS 2–6–3 cells transiently transfected with either LacI-CFP-Nup53 (A, B), LacI-CFP-Nup155 (C, D), or LacI-CFP-Nup58 (E, F) were stained with the indicated antibodies. Chromatin is visualized using DAPI stain. In panels (A) and (B), LacI-CFP-Nup53 was detected by CFP fluorescence. The right hand panels show a merged image of the 3 stains. The smaller insets show a magnified image of the LacO array in each image. Colocalization of the untagged endogenous Nup in question (middle panels) with the transfected LacI-CFP-Nup being tested (left panels) is summarized at the right.
Figure 3.
Recruitment of nucleoporins from other NPC subcomplexes to the LacO array. Immunofluorescence microscopy of U2OS 2–6–3 cells transiently transfected with either LacI-CFP-Nup214 (A) or LacI-CFP-Pom121 (B, C), then stained with antibody against LacI or visualized by CFP fluorescence (left panel). The endogenous nucleoporins Nup133 (A), Nup62 (B), or Nup93 (C) were detected with specific antibodies (middle panel). Chromatin is visualized by DAPI staining. The smaller insets show a magnified image of the LacO array in each image. The right hand panels are a merge of the previous 2 images with DAPI staining. All three cases show examples of positive recruitment of an endogenous nucleoporin from a different subcomplex than that of the LacI-CFP-NupX. Full results are summarized in Table 1.
Figure 4.
Nup133 is able to recruit an extensive array of nucleoporins to the LacO array. Immunofluorescence microscopy was performed on U2OS 2–6–3 cells transiently transfected with LacI-CFP-Nup133. The location of LacI-CFP-Nup133 was visualized by CFP fluorescence (left panel). To assay for recruitment of endogenous nucleoporins, the transfected cells were stained with antibody against: (A) Nup85, (B) Nup160, (C) ELYS, (D) Nup53, (E) Nup93, (F) Nup214, and (G) Nup98, followed by a Texas Red-labeled secondary antibody (middle panel). DNA was visualized by DAPI staining. The smaller panels show a magnified image of the LacO array. The right hand panels are a merge of the previous 2 images together with a DAPI-stained image. We note that when Nup133 was used as the LacI-CFP-tagged fusion protein, a large percentage of the transfected cells showed rim localization of the LacO array. Quantitation is shown in Table 1.
Figure 5.
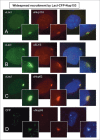
Widespread recruitment of nucleoporins to the LacO array by the LacI-CFP-Nup153 fusion protein. Immunofluorescence microscopy of U2OS 2–6–3 cells transiently transfected with a LacI-CFP-Nup153 construct. The location of LacI-CFP-Nup153 was visualized by CFP fluorescence or staining with anti-LacI antibody (left panel). Endogenous nucleoporin recruitment was assessed with antibody against: (A) Nup133, (B) ELYS, (C) Nup62, or (D) Nup93 (middle panel). Chromatin was visualized by DAPI staining. The smaller panel shows a magnified image of the LacO array for each image; the right hand panel is a merge of the previous 2 images with the DAPI DNA stain. Data for additional nucleoporins tested as well as the quantitation for colocalization and rim localization for all tested nucleoporins are shown in Table 1.
Figure 6.
Nup133 N- and C-terminal protein fragments bind nucleoporins differentially. (A) Full length human Nup133 (1141 aa) and specific mapped domains are shown. The N-terminal, designated Nt-Nup133, consisted of 805 aa and contained the β-propeller domain, the ALPS motif, and a portion of the α-helical domain. The C-terminal Ct-Nup133 construct consisted of aa 917–1141 and contained the site for Nup107 interaction (aa 934–980). (B–C) U20S 2–6–3 cells were transfected with the designated Nup133 fragment. The extent of nucleoporin recruitment was then tested by staining with anti-LacI antibody together with antibodies to Nup160 (B-C) or Pom121, ELYS, Nup53, Nup98, and Nup214 (D). The percentage of transfected cells showing rim localization is shown in last column of (D). The comparative data for full length Nup133 is presented in Table 1.
Figure 7.
The Nup155 R391H mutation that causes human cardiac arrhythmia abrogates binding of its normal nucleoporin partners and rim localization. U2OS 2–6–3 cells were transfected with a LacI-CFP construct of full-length wild type human Nup155 and the R391H mutant version of the same construct. The transfected cells were tested by immunofluorescence for colocalization of Nup53 or Pom121 with the wild type or mutant Nup155. Note that Nup53 and Pom121 were the only tested nucleoporins that had been seen to be recruited to wild type LacI-CFP-Nup155 (see Table 1). Here Nup53 recruitment can be observed with wild type LacI-CFP-Nup155 (A), but not with the R391H mutant Nup155 (B). (C) Quantitation for the level of Nup53 and Pom121 colocalization and rim targeting is shown for the wild type and mutant Nup155 constructs.
Figure 8.
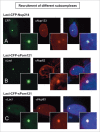
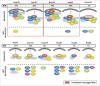
A schematic summary of the recruitment and rim localization results for each LacI-CFP-Nup tested. The individual LacI-tagged Nup construct transfected (outlined in red) is indicated above each drawing. Endogenous nucleoporins tested that were recruited to the LacI-Nup/LacO array are shown above the dotted line, while the non-recruited Nups are shown below the dotted line. Nups belonging to the same subcomplex are in the same color (and correlate with the subcomplexes in Fig. 1A). Rim localization of the LacI-Nup/LacO array (% of cells) is denoted by close proximity of the recruited nucleoporins to the double nuclear membrane lines, and also by the % of cells that showed rim localization of the array (RIM: %). Double headed arrows indicate the Nups where the LacI-Nup/LacO array was found adjacent to the membranes, but in lesser percentages. A number of LacI-Nup/LacO arrays (sPom121, Nup214, Nup58, Nup85, and Nup98) showed essentially no rim localization. Red squares denote the members of the 2 subcomplexes that are able to initiate both extensive Nup recruitment to the array and rim localization of the array.
Similar articles
- Marker gene tethering by nucleoporins affects gene expression in plants.
Smith S, Galinha C, Desset S, Tolmie F, Evans D, Tatout C, Graumann K. Smith S, et al. Nucleus. 2015;6(6):471-8. doi: 10.1080/19491034.2015.1126028. Epub 2015 Dec 10. Nucleus. 2015. PMID: 26652762 Free PMC article. - Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export.
Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. Vasu S, et al. J Cell Biol. 2001 Oct 29;155(3):339-54. doi: 10.1083/jcb.200108007. Epub 2001 Oct 29. J Cell Biol. 2001. PMID: 11684705 Free PMC article. - Structural characterization of altered nucleoporin Nup153 expression in human cells by thin-section electron microscopy.
Duheron V, Chatel G, Sauder U, Oliveri V, Fahrenkrog B. Duheron V, et al. Nucleus. 2014;5(6):601-12. doi: 10.4161/19491034.2014.990853. Nucleus. 2014. PMID: 25485891 Free PMC article. - The Role of Nucleoporin Elys in Nuclear Pore Complex Assembly and Regulation of Genome Architecture.
Shevelyov YY. Shevelyov YY. Int J Mol Sci. 2020 Dec 13;21(24):9475. doi: 10.3390/ijms21249475. Int J Mol Sci. 2020. PMID: 33322130 Free PMC article. Review. - Structure and Assembly of the Nuclear Pore Complex.
Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Hampoelz B, et al. Annu Rev Biophys. 2019 May 6;48:515-536. doi: 10.1146/annurev-biophys-052118-115308. Epub 2019 Apr 3. Annu Rev Biophys. 2019. PMID: 30943044 Review.
Cited by
- WNT signaling and AHCTF1 promote oncogenic MYC expression through super-enhancer-mediated gene gating.
Scholz BA, Sumida N, de Lima CDM, Chachoua I, Martino M, Tzelepis I, Nikoshkov A, Zhao H, Mehmood R, Sifakis EG, Bhartiya D, Göndör A, Ohlsson R. Scholz BA, et al. Nat Genet. 2019 Dec;51(12):1723-1731. doi: 10.1038/s41588-019-0535-3. Epub 2019 Nov 29. Nat Genet. 2019. PMID: 31784729 - Nucleoporin 107, 62 and 153 mediate Kcnq1ot1 imprinted domain regulation in extraembryonic endoderm stem cells.
Sachani SS, Landschoot LS, Zhang L, White CR, MacDonald WA, Golding MC, Mann MRW. Sachani SS, et al. Nat Commun. 2018 Jul 18;9(1):2795. doi: 10.1038/s41467-018-05208-2. Nat Commun. 2018. PMID: 30022050 Free PMC article. - Intranuclear dynamics of the Nup107-160 complex.
Morchoisne-Bolhy S, Geoffroy MC, Bouhlel IB, Alves A, Audugé N, Baudin X, Van Bortle K, Powers MA, Doye V. Morchoisne-Bolhy S, et al. Mol Biol Cell. 2015 Jun 15;26(12):2343-56. doi: 10.1091/mbc.E15-02-0060. Epub 2015 Apr 22. Mol Biol Cell. 2015. PMID: 25904327 Free PMC article. - The Nup2 meiotic-autonomous region relieves inhibition of Nup60 to promote progression of meiosis and sporulation in Saccharomyces cerevisiae.
Komachi K, Burgess SM. Komachi K, et al. Genetics. 2022 May 5;221(1):iyac045. doi: 10.1093/genetics/iyac045. Genetics. 2022. PMID: 35302609 Free PMC article. - Nucleoporin genes in human diseases.
Nofrini V, Di Giacomo D, Mecucci C. Nofrini V, et al. Eur J Hum Genet. 2016 Oct;24(10):1388-95. doi: 10.1038/ejhg.2016.25. Epub 2016 Apr 13. Eur J Hum Genet. 2016. PMID: 27071718 Free PMC article. Review.
References
- Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett 2008; 582:2004-16; PMID:18328825; http://dx.doi.org/10.1016/j.febslet.2008.02.067 - DOI - PubMed
- D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 2008; 18:11; PMID:18786826; http://dx.doi.org/10.1016/j.tcb.2007.11.001 - DOI - PMC - PubMed
- Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol 2005; 15:6; PMID:15837182; http://dx.doi.org/10.1016/j.sbi.2005.03.003 - DOI - PubMed
- Wente SR. Gatekeepers of the nucleus. Science 2000; 288:1374-7; PMID:10827939; http://dx.doi.org/10.1126/science.288.5470.1374 - DOI - PubMed
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell 2005; 17:83-92; PMID:15629719; http://dx.doi.org/10.1016/j.molcel.2004.12.010 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous